Fig. 7.
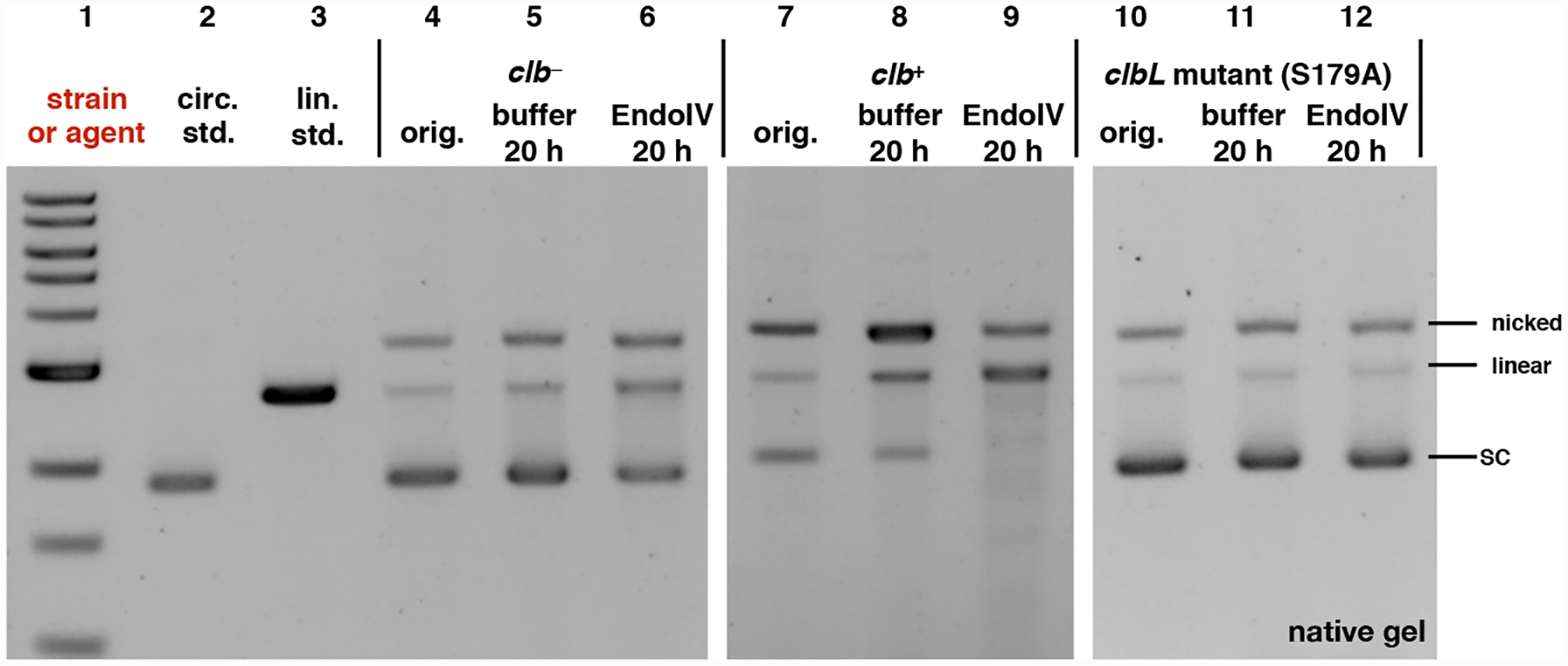
Exposure of pUC19 plasmid DNA to clb+, followed by incubation with Endo IV leads to consumption of undamaged plasmid and formation of nicked and linearized DNA. This is not observed in the clb− or clbL mutant controls. DNA ladder (Lane #1); circular pUC19 DNA standard (Lane #2); linearized pUC19 DNA standard (Lane # 3); circular pUC19 DNA isolated from co-culture with clb− BW25113 E. coli (Lane #4), reacted with buffer (Lane #5), reacted with Endonuclease IV (Lane #6); circular pUC19 DNA isolated from co-culture with clb+ BW25113 E. coli (Lane #7), reacted with buffer (Lane #8), reacted with Endonuclease IV (Lane #9); circular pUC19 DNA isolated from co-culture with clbL mutant (S179A) BW25113 E. coli (Lane #10), reacted with buffer (Lane #11), reacted with Endonuclease IV (Lane #12). Conditions (Lane #4–#6): circular pUC19 DNA from co-culture with clb− BW25113 E. coli in M9-CA media for 4 h at 37 °C (Lane # 4); the DNA (3.9 μM base pair) was reacted with NEBuffer 3.1 (New England Biolabs®), pH 7.9, at 37 °C for 20 hours (Lane #5); the DNA (3.9 μM base pair) was further reacted with 20 units of Endonuclease IV in NEBuffer 3.1® (New England Biolabs®), pH 7.9, at 37 °C for 20 hours (Lane #6). Conditions (Lane #7–#9): circular pUC19 DNA isolated from co-culture with clb+ BW25113 E. coli. in in M9-CA media for 4 h at 37 °C (Lane #7); the DNA (3.9 μM base pair) was reacted with NEBuffer 3.1 (New England Biolabs®), pH 7.9, at 37 °C for 20 hours (Lane #8); the DNA (3.9 μM base pair) was reacted with 20 units of Endonuclease IV in NEBuffer 3.1® (New England Biolabs®), pH 7.9, at 37 °C for 20 hours (Lane #9). Conditions (Lane #10–#12): circular pUC19 DNA isolated from co-culture with clbL mutant (S179A) BW25113 E. coli. in M9-CA media for 4 h at 37 °C (Lane #10); the DNA (3.9 μM base pair) was reacted with NEBuffer 3.1® (New England Biolabs®), pH 7.9, at 37 °C for 20 hours (Lane #11); the DNA (3.9 μM base pair) was reacted with 20 units of Endonuclease IV in NEBuffer 3.1® (New England Biolabs®), pH 7.9, at 37 °C for 20 hours (Lane #12). The DNA was not re-purified and was directly analyzed by native agarose gel electrophoresis (90 V, 1.5 hr).
