Fig. 9.
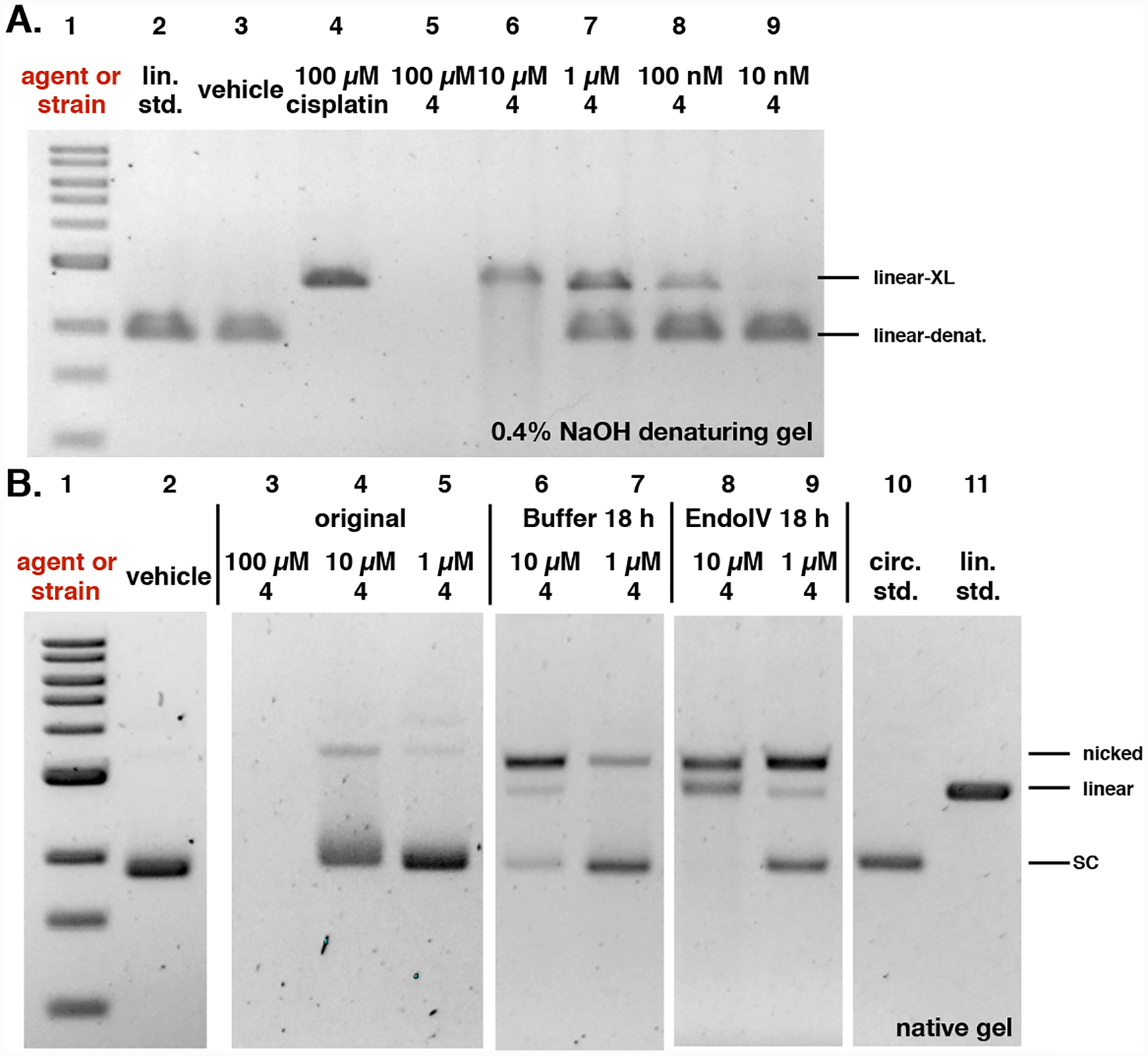
Analysis of induction of AP sites by the colibactin precursor 4. A. Incubation of plasmid pUC19 DNA exposed to 4 in buffer for 18 h results in minor nicking and cleavage. B. Addition of EndoIV increases the amount of nicked and cleaved plasmid. Conditions: A. 5% DMSO was used as vehicle (negative control), and 100 μM cisplatin was used as positive control. DNA ladder (Lane #1); linearized pUC19 DNA standard (Lane #2); 5% DMSO (Lane #3); 100 μM cisplatin (Lane #4); 100 μM 4 (Lane #5); 10 μM 4 (Lane #6); 1 μM 4 (Lane #7); 100 nM 4 (Lane #8); 10 nM 4 (Lane #9). Conditions (Lane #3): linearized pUC19 DNA (15.4 μM in base pairs), 5% DMSO (vehicle), 10 mM citric buffer, pH 5.0, 4 h, 37 °C. Conditions (Lane #4): linearized pUC19 DNA (15.4 μM in base pairs), 5% DMSO (vehicle), 100 μM cisplatin, 10 mM citric buffer, pH 5.0, 4 h, 37 °C. Conditions (Lanes #5–#9): circular pUC19 DNA (15.4 μM in base pairs), 4 (100 μM–10 nM), 5% DMSO, 10 mM citric buffer, pH 5.0, 4 h, 37 °C. The DNA was analyzed by 0.4% NaOH denaturing agarose gel electrophoresis (90 V, 1.5 h). B. 5% DMSO was used as vehicle. DNA ladder (Lane #1); 5% DMSO (Lane #2); 100 μM 4 (Lane #3); 10 μM 4 (Lane #4); 1 μM 4 (Lane #5); post buffer-reacted after 10 μM 4 (Lane #6); post buffer-reacted after 1 μM 4 (Lane #7); post EndoIV-reacted after 10 μM 4 (Lane #8); post EndoIV-reacted after 1 μM 4 (Lane #9); circular pUC19 plasmid standard (Lane #10); linearized pUC19 plasmid standard (Lane #11). Conditions (Lane #2): circular pUC19 DNA (15.4 μM in base pairs), 5% DMSO (vehicle), 10 mM citric buffer, pH 5.0, 4 h, 37 °C. Conditions (Lanes #3–#5): circular pUC19 DNA (15.4 μM in base pairs), 4 (100 μM–1 μM), 5% DMSO, 10 mM citric buffer, pH 5.0, 3 h, 37 °C. Conditions (Lanes #6–#7): 4 (10 μM–1 μM)-treated circular pUC19 DNA (3.9 μM in base pairs), NEBuffer 3.1® (New England Biolabs®), pH 7.9, at 37 °C for 18 h. Conditions (Lanes #8–#9): 4 (10 μM–1 μM)-treated circular pUC19 DNA (3.9 μM in base pairs), 20 units of Endonuclease IV (New England Biolabs®), NEBuffer 3.1® (New England Biolabs®), pH 7.9, at 37 °C for 18 h. The DNA was analyzed by native agarose gel electrophoresis (90 V, 2 h). Linear-denat. = DSB/linearized DNA in denaturing form, linear-XL = DSB/linerized DNA cross-linked by colibactin or cisplatin, SC = supercoiled, nicked = SSB, linear = DSB.
