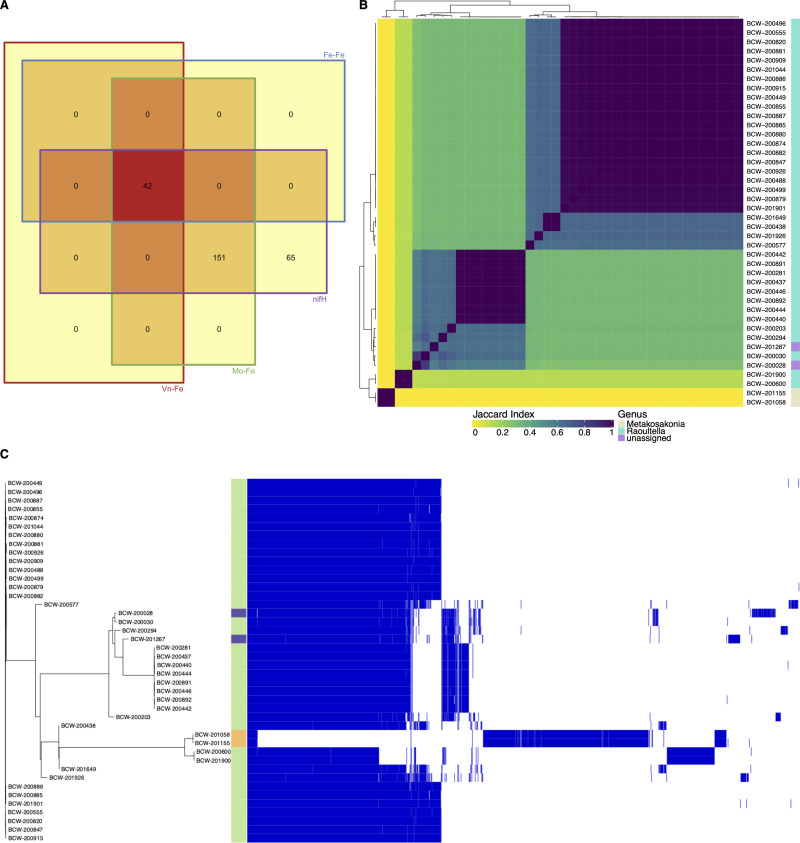
Fig 5. Mucilage bacterial isolates exhibit alternative nitrogenase genes.
A) The presence of predicted protein sequences in diazotrophic isolate genomes was detected using TIGRFAM HMMs corresponding to the Fe-Fe and Vn-Fe alternative nif genes (anfD, anfK, anfG, vnfD, vnfK, vnfG) along with HMMs for nifHDK. Genomes with detected presence of the targeted genes were compared and quantified using a Venn Diagram to determine the list of diazotrophs with genes resembling Vn-Fe, Fe-Only, Mo-Fe Type nitrogenases and nifH. B) Genomes with alternative nif genes were compared using Sourmash [19] and visualized as a composite matrix that included annotation of genus level classification. C) Pangenome analysis of diazotrophs with alternative nif genes was conducted using Roary [36] and data for gene presence and absence was visualized using Phandango [37] along with genus classification data from Sourmash LCA. Orange annotations indicate genomes classified as Metakosakonia, green annotations indicate Raoultella isolates, and purple annotations indicate “unassigned” classification at the genus level.