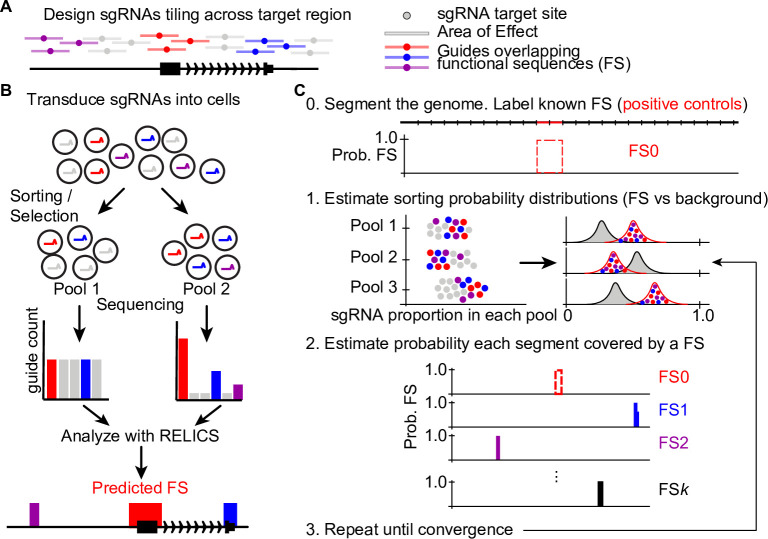
Fig 1. Schematic of a tiling CRISPR screen and RELICS.
(A) sgRNAs are designed to target sites tiling across a genomic region. Bars around each target site indicate the area of effect. sgRNAs ‘overlap’ genome sequences within their area of effect. In this example, sgRNAs overlapping functional sequences (FSs) are colored red, purple and blue. (B) In a CRISPR screen, sgRNAs are introduced into cells and the cells are sorted into pools or subjected to selection (e.g. for proliferation or survival). The sgRNAs in each pool are counted and analyzed by RELICS to predict FSs. (C) RELICS predicts FSs using an Iterative Bayesian Stepwise Selection Algorithm. Step 0 (initialization): the screened region is divided into small (e.g. 100bp) genome segments; segments containing known FSs are labeled FS0. Step 1: Using the predicted FSs, RELICS estimates sorting probability distributions for sgRNAs that do/do-not overlap FSs. Step 2: RELICS uses the sorting probability distributions to compute the probability each genome segment contains a functional sequence. Step 3: Steps 1–3 are repeated until convergence.