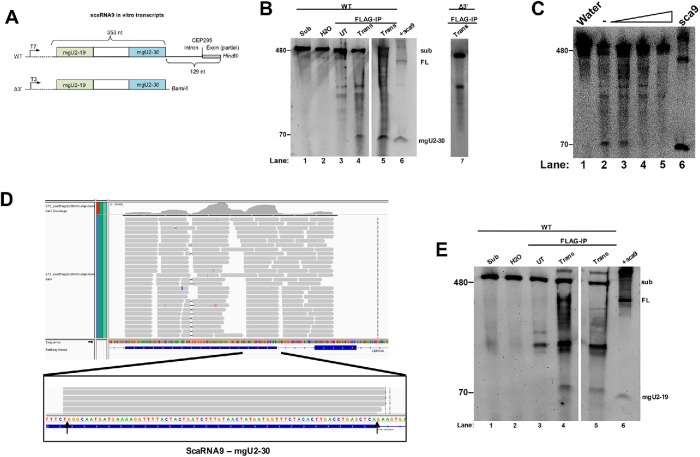
FIGURE 4:
In vitro processing of primary-scaRNA9 by Drosha/DGCR8. (A) WT and Δ3’ scaRNA in vitro transcribed substrates. (B) Northern blot of RNA isolated from the in vitro Drosha/DGCR8 processing assays. FL scaRNA9 and the mgU2-30 fragment were detected with a probe that hybridizes to the mgU2-30 region. Substrate was incubated with FLAG beads from Drosha/FLAG-DGCR8 transfected lysate (Trans, lanes 4, 5, and 7). Control reactions include substrate (lane 1), substrate incubated with water alone (lane 2), substrate incubated with FLAG beads from nontransfected cell lysate (lane 3), and a positive control of ectopically expressed scaRNA9 RNA (lane 6). (C) Processing assay with Drosha/DGCR8 FLAG beads using WT scaRNA9 substrate and increasing amounts of salt (lanes 3–5). Ectopically expressed scaRNA9 RNA is shown in lane 6. (D) RNA sequencing reads of in vitro Drosha processed scaRNA9. Arrows indicate the 5′ and 3′ ends of mgU2-30 as described previously (Tycowski et al., 2004). (E) Northern blot of RNA generated from in vitro Drosha processing assay and probed with a scaRNA9 mgU2-19 probe. A band around 79 nts appears in lanes 4 and 5, consistent with the mgU2-19 fragment. This gel was organized as described in B.