ABSTRACT
Integrin function depends on the continuous internalization of integrins and their subsequent endosomal recycling to the plasma membrane to drive adhesion dynamics, cell migration and invasion. Here we assign a pivotal role for Rabgap1 (GAPCenA) in the recycling of endocytosed active β1 integrins to the plasma membrane. The phosphotyrosine-binding (PTB) domain of Rabgap1 binds to the membrane-proximal NPxY motif in the cytoplasmic domain of β1 integrin subunits on endosomes. Silencing Rabgap1 in mouse fibroblasts leads to the intracellular accumulation of active β1 integrins, alters focal adhesion formation, and decreases cell migration and cancer cell invasion. Functionally, Rabgap1 facilitates active β1 integrin recycling to the plasma membrane through attenuation of Rab11 activity. Taken together, our results identify Rabgap1 as an important factor for conformation-specific integrin trafficking and define the role of Rabgap1 in β1-integrin-mediated cell migration in mouse fibroblasts and breast cancer cells.
KEY WORDS: Rabgap1, Integrin recycling, Active β1 integrins, Cell migration, Cancer cell invasion
Summary: The β1 integrin interactor Rabgap1 promotes recycling of active β1 integrins from endosomes to enhance cell migration on 2D surfaces as well as cell invasion into a 3D matrix.
INTRODUCTION
Essentially all morphogenetic events of multicellular organisms, tissue homeostasis, wound healing and tumorigenesis require dynamic cell–cell and cell–matrix adhesion. Integrins are heterodimeric transmembrane receptors for extracellular matrix (ECM) proteins and cell counter receptors that link the extracellular environment to the intracellular actomyosin cytoskeleton. Hence, integrins fulfil key functions in cell adhesion and migration crucial for development, physiology and pathology (Hynes, 2002). Integrin function is regulated on different levels, including activation (which causes conformation changes and ligand binding), clustering, the assembly of a signaling hub where biochemical and biophysical signals converge, and trafficking through the endosomal system (Moreno-Layseca et al., 2019; Moser et al., 2009; Sun et al., 2019).
Integrin activation is induced by talin and kindlin (also known as fermitin) proteins binding to the NxxY motifs of β integrin tails (Böttcher et al., 2017; Calderwood et al., 2013; Moser et al., 2009; Theodosiou et al., 2016) and is characterized by the unbending of the integrin ectodomain, the unclasping of the transmembrane and cytoplasmic domains and the swing-out of the β hybrid domain. These distinct conformations exhibit different ligand-binding affinities, and therefore the shift from the bent-closed conformation to the extended-open conformation is termed ‘integrin activation’ (Li and Springer, 2017; Zhu et al., 2013). Impaired integrin activation is implicated in multiple pathological conditions (Hegde and Raghavan, 2013; Pozzi and Zent, 2013; Winograd-Katz et al., 2014).
Although the prime function of integrins is the recruitment of adaptor and signaling proteins at the plasma membrane upon activation and ligand binding, integrins continuously cycle between the cell surface and internal compartments. After a residence time of ∼10–30 min, integrins are internalized via clathrin-dependent and -independent mechanisms and routed into the endosomal network, where they are recycled (re-exocytosed) back to the plasma membrane to be reused at adhesion sites, or sorted to late endosomes and lysosomes for degradation (Caswell et al., 2009; Moreno-Layseca et al., 2019). The constant integrin endo/exocytosis cycle is crucial for cell migration (Moreno-Layseca et al., 2019; Shafaq-Zadah et al., 2016; White et al., 2007) and cancer cell invasion (Caswell et al., 2008; Muller et al., 2009; Paul et al., 2015) because it regulates integrin distribution on the cell surface and assists in the formation and disassembly of cell–matrix adhesions, critical determinants for migration speed and persistence during directional migration. Interestingly, integrin trafficking is not only regulated by integrin-tail-binding proteins, including Rab-coupling protein (also known as RAB11FIP1), Rab21, PPFIA1, SNX17 and SNX31 (Böttcher et al., 2012; Caswell et al., 2008; Mana et al., 2016; Pellinen et al., 2006; Steinberg et al., 2012; Tseng et al., 2014), but also by the integrin conformation itself (Arjonen et al., 2012; Valdembri et al., 2009). Active β1 integrins have been detected inside endosomes (Alanko et al., 2015; Rainero et al., 2015), and active unligated β1 integrins traffic from talin- and FAK-positive endosomes to the plasma membrane (Nader et al., 2016). Whereas inactive β1 integrins are trafficked via a Rab4-dependent short-loop recycling pathway or undergo retrograde trafficking to the trans-Golgi network, active β1 integrins recycle through the Rab11-dependent long-loop pathway of the perinuclear recycling compartment or post-Golgi carriers (Arjonen et al., 2012; Mana et al., 2016; Powelka et al., 2004; Shafaq-Zadah et al., 2016). However, the molecular mechanisms underlying the conformation-specific recycling of β1 integrins and their connection to specific endosomal Rab GTPase-mediated trafficking routes are only poorly understood.
Rabgap1, also known as GAPCenA, was originally described as a Rab6 GTPase-activating protein (GAP) (Cuif et al., 1999). Although the GTPase function for Rab6 is debated, Rabgap1 was shown to bind Rab4, Rab11 and Rab36 and promote their GTPase activity (Cuif et al., 1999; Fuchs et al., 2007; Kanno et al., 2010). Rabgap1 consists of an N-terminal phosphotyrosine-binding (PTB) domain, a conserved catalytic TBC (TRE2/BUB2/CDC16) (Frasa et al., 2012) domain in the central part of the molecule that promotes GTP hydrolysis, and a C-terminal coiled-coil (CC) domain. Rabgap1 function has been linked to Golgi apparatus dynamics (Cuif et al., 1999; Kanno et al., 2010), the modulation of intracellular lysosomal and early endosomal positioning and vesicular trafficking in a GAP-dependent manner (Kawasaki et al., 2018).
Here we identified Rabgap1 as interactor of the proximal NPxY motif of the β1 integrin cytoplasmic tail that promotes recycling of active β1 integrins to enhance cell migration on 2D surfaces as well as cell invasion into a 3D matrix.
RESULTS
Rabgap1 interacts with the proximal NPxY motif of the β1 integrin cytoplasmic tail
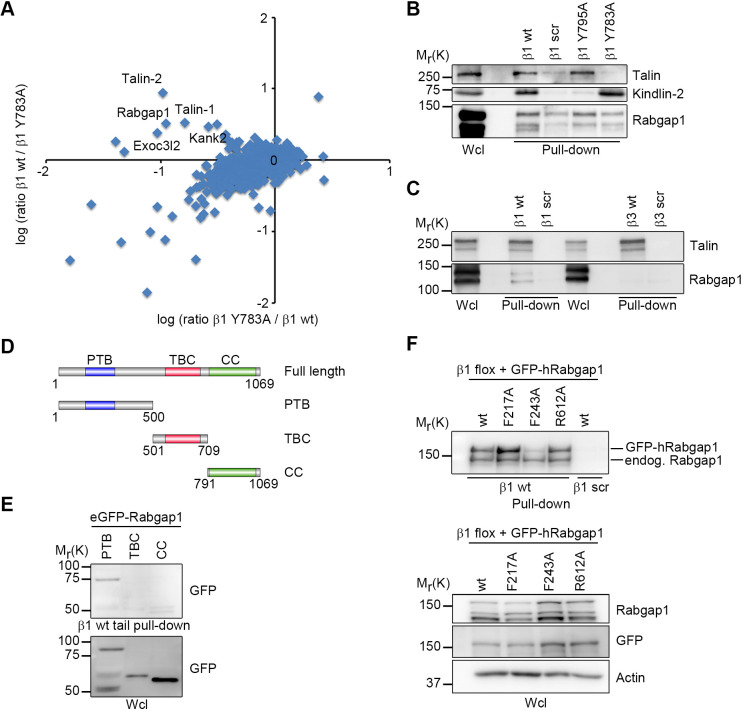
Integrins require proteins binding to their cytosolic domains to regulate their activation, connection to the actin cytoskeleton, signaling and endosomal trafficking. We previously used stable isotope labeling by amino acids in cell culture (SILAC)-based quantitative proteomics in combination with β1 integrin tail peptide pulldown experiments to identify cytosolic integrin interactors in different cell types (Böttcher et al., 2012; Meves et al., 2011, 2013; Schiller et al., 2013). Here, we hypothesized that the membrane-proximal NPxY motif required for integrin activation (Meves et al., 2013; Wegener et al., 2007; Ye et al., 2010) might also play a role in the interaction of proteins involved in conformation-specific endosomal trafficking of β1 integrins. Therefore, SILAC-based quantitative proteomics was used to compare proteins interacting with the full-length mouse β1 wild-type tail peptide (β1 wt) to proteins interacting with a tail peptide harboring a tyrosine to alanine mutation at amino acid 783 (β1 Y783A) in the membrane-proximal NPxY motif (Fig. 1A). Among proteins with high light:heavy isotope ratios, indicative of specific binding to the Y783 residue, were known β1-tail-interacting proteins such as talin-1, talin-2, the talin interactor Kank2 (Sun et al., 2016), exocyst complex component 3 like 2 (Exoc3l2) and Rabgap1 (Fig. 1A). Rabgap1 (also known as TBC1D11 or GAPCenA) belongs to the group of TBC domain proteins and stimulates the GTPase activity of several Rab proteins, including Rab4, Rab11 and Rab36 (Fuchs et al., 2007; Kanno et al., 2010). The physiological functions of Rabgap1 are poorly understood. To confirm the specific interaction between Rabgap1 and Y783 in the β1 integrin cytoplasmic domain, we performed peptide pulldowns with β1 wt, a scrambled peptide and peptides bearing Y783A or Y795A mutations in the NPxY motifs, which inhibit talin and kindlin binding, respectively (Fig. 1B). Consistent with the proteomic data, Rabgap1 was readily detected in peptide pulldowns with β1 wt and β1 Y795A but not with β1 Y783A or scrambled peptides. Rabgap1 is not a general interactor of all β integrin tails, because we observed no binding to β3 integrin tail peptides (Fig. 1C). We verified the binding of Rabgap1 to the membrane-proximal NPxY motif in proximity biotinylation assays using BioID (Kim et al., 2016). Expression of the integrin α5–BioID2 fusion protein, which catalyzes the biotinylation of proteins in proximity to the α5β1 integrin, allowed the pulldown of Rabgap1 in β1 wt- but not in β1 Y783A-expressing mouse fibroblasts (Fig. S1A). Next, to identify the Rabgap1 domain important for β integrin binding, we performed β1 wt peptide pulldowns with cells expressing eGFP-tagged variants of the Rabgap1 PTB, TBC, and CC domains, and found that β1 integrin specifically interacted with the PTB domain of Rabgap1 (Fig. 1D,E). Finally, we expressed GFP-tagged human Rabgap1 variants harboring point mutations in amino acids of the PTB domain predicted to be involved in NPxY motif recognition and coordination (Uhlik et al., 2005), to verify the importance of the PTB domain for integrin binding. Substitution of F243, but not F217, of human Rabgap1 with alanine strongly reduced its ability to bind β1 integrin tail peptides (Fig. 1F). In contrast, abrogating the GAP activity of Rabgap1, by introducing an R612A mutation to abolish the IxxDxxR arginine finger motif (Pan et al., 2006), did not affect β1 integrin binding (Fig. 1F). Taken together, the data demonstrate that Rabgap1 binds the proximal NPxY motif of the β1 integrin cytoplasmic tail through its PTB domain.
Fig. 1.
Rabgap1 interacts with the membrane-proximal NPxY motif of the β1 integrin tail. (A) Scatter plot of β1 wt tail peptide versus β1 Y783A peptide pulldown results. The log2 SILAC ratio of proteins identified with at least two unique peptides in each mass spectrometry run is plotted for the forward pulldown (x axis) and the cross-over pulldown (y axis). Specific interaction partners show inverse ratios between forward and cross-over experiments, grouping them into the upper left quadrant. (B) Western blot confirmation of the mass spectrometry analysis for talin, kindlin-2 and Rabgap1 using the biotinylated integrin β1 peptides indicated (β1 scr, scrambled control peptide). Mutation of the membrane-proximal NPxY motif of the β1 integrin cytoplasmic tail (β1 Y783A) abolished Rabgap1 binding, but mutation of the membrane-distal NxxY motif (β1 Y795A) did not. (C) Western blot analysis of streptavidin-bead pulldown assays with wild-type or scrambled biotinylated β1- and β3-integrin cytoplasmic tail peptides. (D) Schematic representation of the domain structure of human Rabgap1. Rabgap1 contains a PTB domain in the N-terminal region (blue box), a TBC domain in the central region (red box), and a potential CC domain (green box) in the C-terminal region. The three truncated mutants of Rabgap1, PTB (amino acids 1–500), TBC (amino acids 501–790) and CC (amino acids 791–1069) used in the analysis are shown. (E) Western blot analysis of streptavidin-bead pulldown assays using β1 wild-type tail peptides with cell lysates from cells expressing the indicated GFP-tagged Rabgap1 variants. Wild-type β1 integrin tail peptide interacts with the Rabgap1 PTB domain (amino acids 1–500) but not the TBC domain (amino acids 501–790) or CC domain (amino acids 791–1069). (F) Western blot analysis of streptavidin-bead pulldown assays using the wild-type or scrambled biotinylated β1 integrin tail peptides with cell lysates from cells expressing the indicated GFP-tagged human Rabgap1 mutants. Endogenous Rabgap1 is pulled down with wild-type β1 tail peptide, whereas GFP-tagged Rabgap1 F243A fails to bind. Actin is shown as a loading control. CC, terminal coiled-coil domain; endog., endogenous; PTB, phosphotyrosine-binding domain; TBC, TRE2/BUB2/CDC16 domain; Wcl, whole cell lysate.
Rabgap1 colocalizes with integrins in early and recycling endosomes
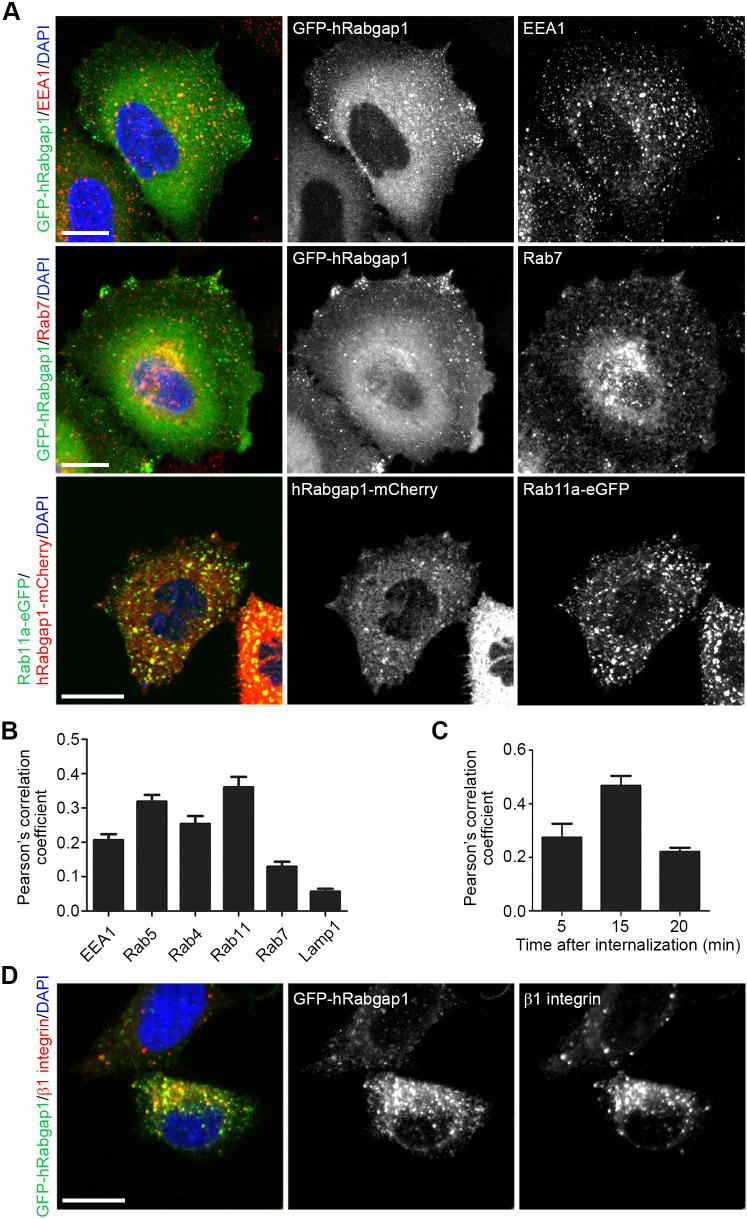
A pool of Rabgap1 is associated with the midbody ring during cytokinesis (Gromley et al., 2005; Miserey-Lenkei et al., 2006) but its subcellular localization in interphase cells is less well characterized. Rabgap1 was mostly cytosolic, but we also observed vesicular staining of GFP- and mCherry-tagged human Rabgap1 in mouse fibroblasts and decided to identify the Rabgap1-positive structures by co-staining with markers for different endosomal compartments (Fig. 2A,B; Fig. S1B,C). Whereas Rabgap1 overlapped with the early endosomal marker Rab5 and with Rab11a-positive recycling endosomes, and partially overlapped with the early endosomal markers EEA1 and Rab4, we did not observe colocalization with the late endosome marker Rab7 and the lysosome marker Lamp1 (Fig. 2A,B; Fig. S1C). To investigate the role of Rabgap1 in integrin trafficking, we checked for colocalization with β1 integrin in endosomes. We labeled cell surface β1 integrins with antibodies prior to internalization on fibroblasts expressing eGFP-tagged Rabgap1 and monitored their trafficking for 5, 15 and 20 min (Fig. 2C,D). Subsequent colocalization quantification revealed a partial overlap of Rabgap1 with β1 integrins 15 min after internalization (Fig. 2C,D). These findings show that a pool of Rabgap1 colocalizes with β1 integrin in early and recycling endosomes.
Fig. 2.
Rabgap1 localizes to early and recycling endosomes in fibroblasts. (A) Confocal images of fibroblasts expressing hRabgap1–mCherry or GFP–hRabgap1 together with eGFP-tagged Rab11a or co-stained with antibodies against EEA1 or Rab7. DAPI (blue) was used to stain nuclei. (B) Colocalization analysis of GFP–hRabgap1 or hRabgap1–mCherry with Rab5–eGFP, Rab4–eGFP, Rab11a–eGFP, Rab7–RFP, Lamp1–RFP or endogenous EEA1. The Pearson's colocalization (PC) coefficient was determined in a region of interest (ROI) inside the cell using the Coloc2 plugin in ImageJ (mean+s.e.m., n=15). (C) Quantification of β1 integrin and GFP–hRabgap1 colocalization after surface labeling with a total anti-β1 integrin antibody and internalization for 5, 15 and 20 min at 37°C. The PC coefficient was measured in a ROI inside the cell using the Coloc2 plugin in ImageJ (mean+s.e.m., n=15). (D) Localization of endogenous β1 integrin after surface labeling with a total anti-β1 integrin antibody and internalization for 15 min at 37°C in GFP–hRabgap1- expressing cells. DAPI (blue) was used to stain nuclei. Scale bars: 10 μm.
Rabgap1 promotes recycling of active β1 integrins
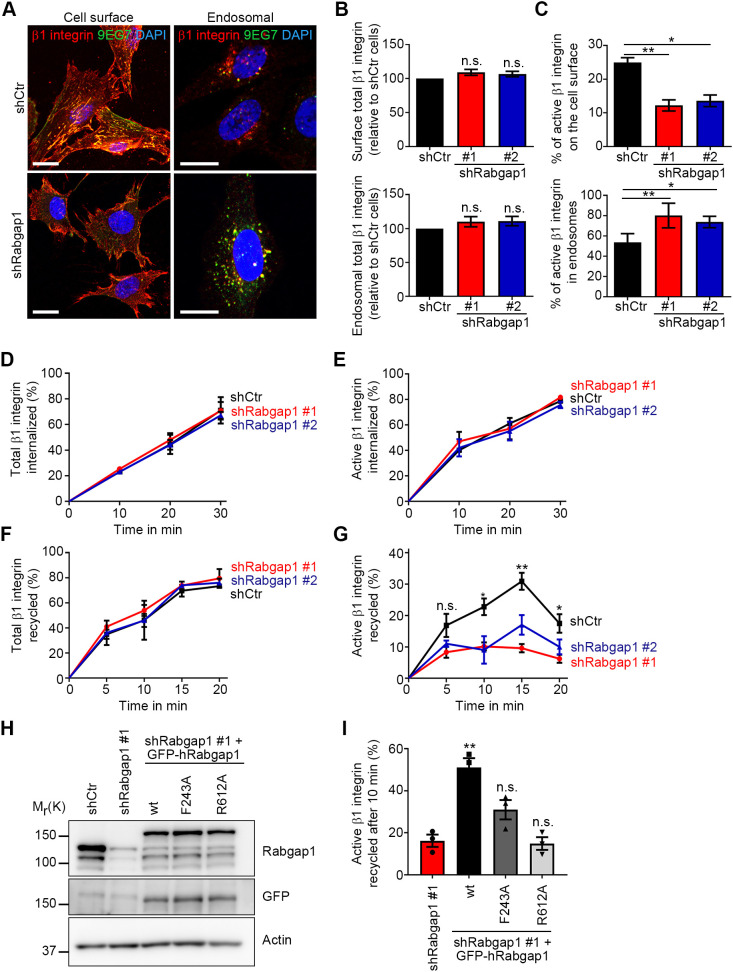
To determine whether Rabgap1 regulates β1 integrin trafficking, we depleted Rabgap1 in mouse fibroblasts by retroviral expression of short hairpin RNA (shRNA; shRabgap1#1, shRabgap1#2) (Fig. S2A) and monitored β1 integrin trafficking using an antibody-based assay (Powelka et al., 2004). Total and active β1 integrins were labeled on the cell surface of control or Rabgap1-depleted cells by different antibodies [using 9EG7 to detect extended β1 integrin ectodomain conformations (Bazzoni et al., 1995; Su et al., 2016)], and the itineraries were followed along the endocytic pathway (Fig. 3A). Rabgap1 depletion affected neither the cell surface level of the total β1 and β3 integrins (Fig. 3A,B; Fig. S2C) nor the amount of total β1 integrin in endosomal vesicles after 30 min of endocytosis (Fig. 3A,B). Interestingly, however, loss of Rabgap1 was accompanied by a significant reduction in the cell-surface levels of the active β1 integrin and an increased intracellular accumulation of the active receptor after internalization (Fig. 3A,C), suggesting selective impairment in trafficking of the active β1 integrin in the absence of Rabgap1. In control cells, we observed that serum induced the recycling of endocytosed total and active β1 integrins back to the plasma membrane, whereas Rabgap1-depleted serum-treated cells failed to efficiently recycle active β1 integrins, which were retained in the early endosomal EEA1-positive compartment (Fig. S2D). To extend these findings, we determined the internalization and recycling kinetics of β1 integrins by surface labeling of β1 integrin in control and Rabgap1-depleted cells with cleavable biotin, followed by capture ELISA (Roberts et al., 2001). Whereas total and active β1 integrins internalized with the same kinetics in all cell lines (Fig. 3D,E), the recycling of active but not total β1 integrin to the plasma membrane was strongly reduced in Rabgap1-depleted fibroblasts (Fig. 3F,G). To test whether Rabgap1 GAP activity or its ability to interact with β1 integrin are important for mediating active β1 integrin recycling, we expressed shRNA-resistant GFP-tagged wild-type (wt), integrin-binding-deficient (F243A) or GAP-activity-deficient (R612A) human Rabgap1 in Rabgap1-depleted fibroblasts (Fig. 3H) and determined the recycling rates after surface biotinylation. Whereas wild-type Rabgap1 increased the recycling of active β1 integrin, expression of Rabgap1 F243A or Rabgap1 R621A did not rescue the recycling defect (Fig. 3I). Taken together, these data indicate that the β1 integrin–Rabgap1 interaction and Rabgap1 GAP activity are required for efficient recycling of active β1 integrin from early endosomes.
Fig. 3.
Rabgap1 promotes recycling of extended β1 integrins. (A) Confocal images of control (shCtr) and Rabgap1-depleted fibroblasts (shRabgap1) double labeled with antibodies against total (red) and active (9EG7, green) β1 integrin at 4°C for 30 min. The cells were either directly fixed (cell surface) or integrin internalization was induced at 37°C for 30 min followed by a low pH wash at room temperature to remove the antibody remaining at the cell surface (endosomal). DAPI (blue) was used to stain nuclei. Scale bars: 10 μm. (B,C) Quantification of total (B) and active (C) β1 integrin levels on the cell surface or in endosomes by their fluorescence signal intensity in control (shCtr) and Rabgap1-depleted (shRabgap1#1, shRabgap#2) cells. The fluorescence signal intensities of internalized β1 integrin were measured from confocal images using a region of interest drawn inside the cell with the plasma membrane as the boundary. Integrated densities were measured using ImageJ, and the internalized signals were normalized against the total fluorescence signal of the same slice. Data are mean±s.e.m. of n=30 cells. *P<0.05; **P<0.01; n.s., not significant (unpaired t-test). (D,E) Quantification of total (D) and active (E) β1 integrin internalization in control (shCtr) or Rabgap1-depleted fibroblasts (shRabgap1#1, shRabgap#2) by capture ELISA. Data are mean±s.e.m., n=3. (F,G) Quantification of total (F) and active (G) β1 integrin recycling in control (shCtr) or Rabgap1-depleted cells (shRabgap1#1, shRabgap#2) by capture ELISA. Data are mean±s.e.m., n=3. *P<0.05; **P<0.01; n.s., not significant (unpaired t-test). (H) Western blot of control (shCtr), Rabgap1-depleted (shRabgap1#1) and Rabgap1-depleted fibroblasts after re-expression of wild-type or mutant GFP-tagged human Rabgap1 variants. Actin serves as loading control. (I) Quantification of active β1 integrin recycling after 10 min of recycling in Rabgap1-depleted and rescue cell lines after re-expression of wild-type or mutant GFP-tagged human Rabgap1 variants, as assayed by capture ELISA. Data are mean±s.e.m., n=3. **P<0.01; n.s., not significant (unpaired t-test).
Regulation of Rab11 activity by Rabgap1 is critical for β1 integrin recycling
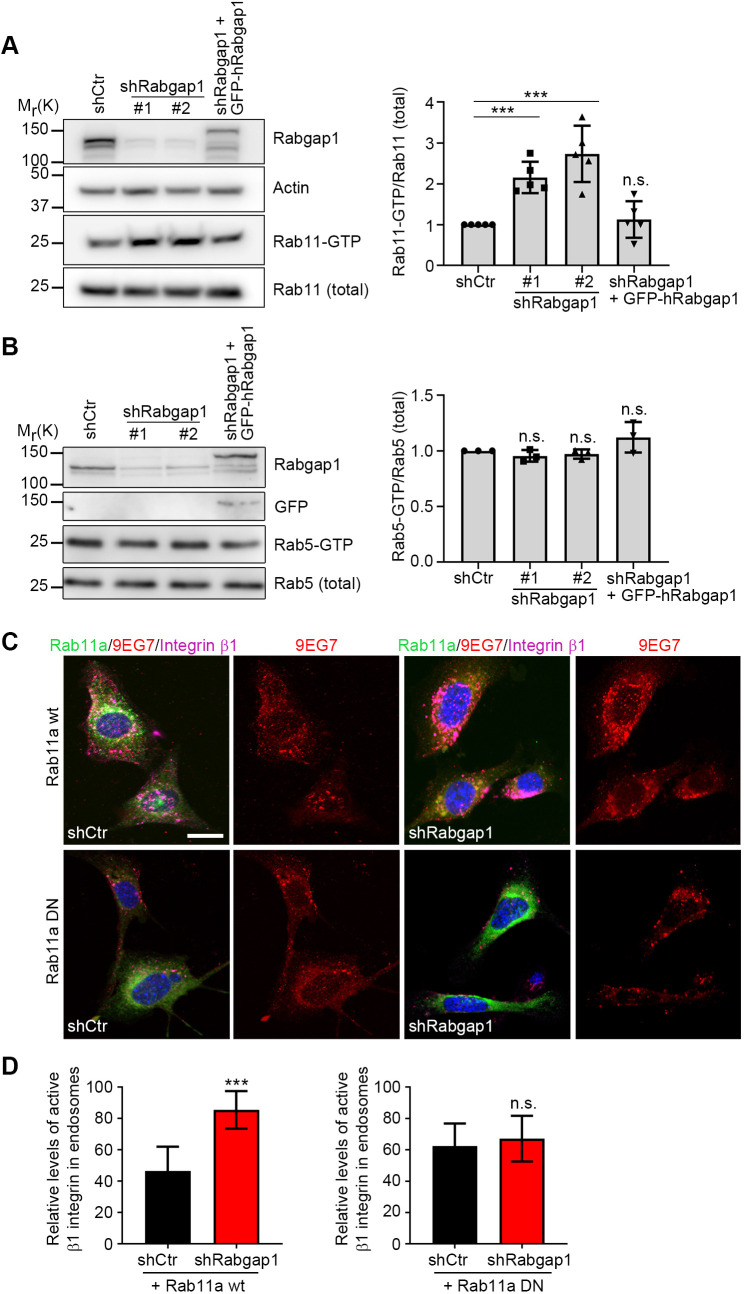
Spatio-temporal regulation of Rab and Arf GTPase activities is critical for controlling the endosomal recycling of α5β1 integrins (Caswell et al., 2008; Pellinen et al., 2006; Qu et al., 2016; Sahgal et al., 2019; Shafaq-Zadah et al., 2016). Because Rabgap1 regulates the GTPase activity of several Rab proteins (Fuchs et al., 2007; Kanno et al., 2010), we analyzed Rab4, Rab11 and Rab36 activity in control and Rabgap1-depleted mouse fibroblasts. Rab36 function has been linked to the spatial distribution of late endosomes and lysosomes (Chen et al., 2010; Matsui et al., 2012), and Rab4a overexpression produces EEA1-positive enlarged endosomes in MCF10A cells that display prolonged and amplified epidermal growth factor (EGF)-induced EGF receptor activation (Tubbesing et al., 2020). We did not observe changes in the spatial distribution of late endosomes or lysosomes in Rabgap1-depleted fibroblasts, indicating that Rab36 activity is not severely affected in these cells (Fig. S3A). Although the size and distribution of EEA1-positive endosomes was also unaffected (Fig. S3A), we observed a prolonged EGF receptor activation in Rabgap1-depleted cells, which was rescued by re-expression of wild-type GFP-tagged human Rabgap1 (Fig. S3B) and could be an indication of increased Rab4a activity in Rabgap1-depleted cells. Finally, we analyzed Rab11 activity in control and Rabgap1-depleted fibroblasts by pulling down GTP-bound, active Rab11 using a C-terminal fragment of the Rab11 effector FIP3 (also known as RAB11FIP3), which specifically binds to the active GTP-bound form of Rab11 (Eathiraj et al., 2006). We found a 2.5-fold Rab11 hyper-activation in Rabgap1-depleted cells (Fig. 4A), whereas the amount of activated Rab5 was unaffected (Fig. 4B). Expression of shRNA-resistant GFP-tagged human Rabgap1 in Rabgap1-depleted cells restored GTP-bound Rab11 to control levels (Fig. 4A).
Fig. 4.
Rab11 drives the recycling of active β1 integrin to the plasma membrane. (A) Western blot and densitometric analysis of Rab11 activation in control (shCtr), Rabgap1-depleted (shRabgap1#1, shRabgap1#2) and GFP-tagged human Rabgap1-expressing rescue (shRabgap1+ GFP–hRabgap1) fibroblasts. The bar chart shows Rab11 activation normalized against total Rab11 levels. Data are mean±s.d. n=5. ***P<0.001; n.s., not significant (unpaired t-test). (B) Western blot and densitometric analysis of Rab5 activation in control (shCtr), Rabgap1-depleted (shRabgap1#1, shRabgap1#2) and GFP-tagged human Rabgap1-expressing rescue (shRabgap1+GFP–hRabgap1) fibroblasts. The bar chart shows Rab5 activation normalized against total Rab5 levels. Data are mean±s.d. n=3. n.s., not significant (unpaired t-test). (C) Confocal microscopy images of internalized total (magenta) and active (9EG7, red) β1 integrin in control (shCtr) or Rabgap1-depleted fibroblasts (shRabgap1) expressing either the GFP-tagged wild-type (Rab11a wt) or dominant-negative Rab11a (Rab11a DN). β1 integrins were labeled on the cell surface at 4°C for 30 min then internalized at 37°C for 30 min before removing the remaining antibodies on the cell surface by an acid wash. Scale bar: 10 μm. (D) Quantification of fluorescence signal intensities of internalized 9EG7-positive, active β1 integrins in the indicated cell lines from confocal images. A region of interest drawn inside the cell with plasma membrane as the boundary was used to measure internalized signal. Integrated densities were measured using ImageJ, and the internalized signals were normalized against the total fluorescence signal of the same slice. Data are mean±s.d. of n=30 cells. ***P<0.001; n.s., not significant (unpaired t-test).
Because active α5β1 integrin traffics predominantly along the Rab11-dependent long-loop pathway (Arjonen et al., 2012), we tested whether Rab11 activity is responsible for α5β1 integrin recycling downstream of Rabgap1. For this, we monitored total and active β1 integrin trafficking with the antibody-based assay in control and Rabgap1-depleted fibroblasts expressing either wild-type (Rab11a wt) or a dominant-negative variant of human Rab11a (S25N; Rab11a DN). At 30 min after internalization, Rabgap1-depleted cells expressing Rab11a wt exhibited a higher amount of active β1 integrin (Fig. 4C,D) in endosomes compared to that in control cells (46% in control cells, 86% in Rabgap1-depleted cells), confirming the previous results. In contrast, we observed a rescue in the level of intracellular active β1 integrin in Rabgap1-depleted fibroblasts expressing dominant-negative Rab11a (62% in control cells, 67% in Rabgap1-depleted cells) (Fig. 4C,D). Overall, these data show that hyper-activation of Rab11 after Rabgap1 depletion disrupts proper recycling of active β1 integrins to the plasma membrane.
Rabgap1 is a regulator of β1-integrin-mediated cell functions
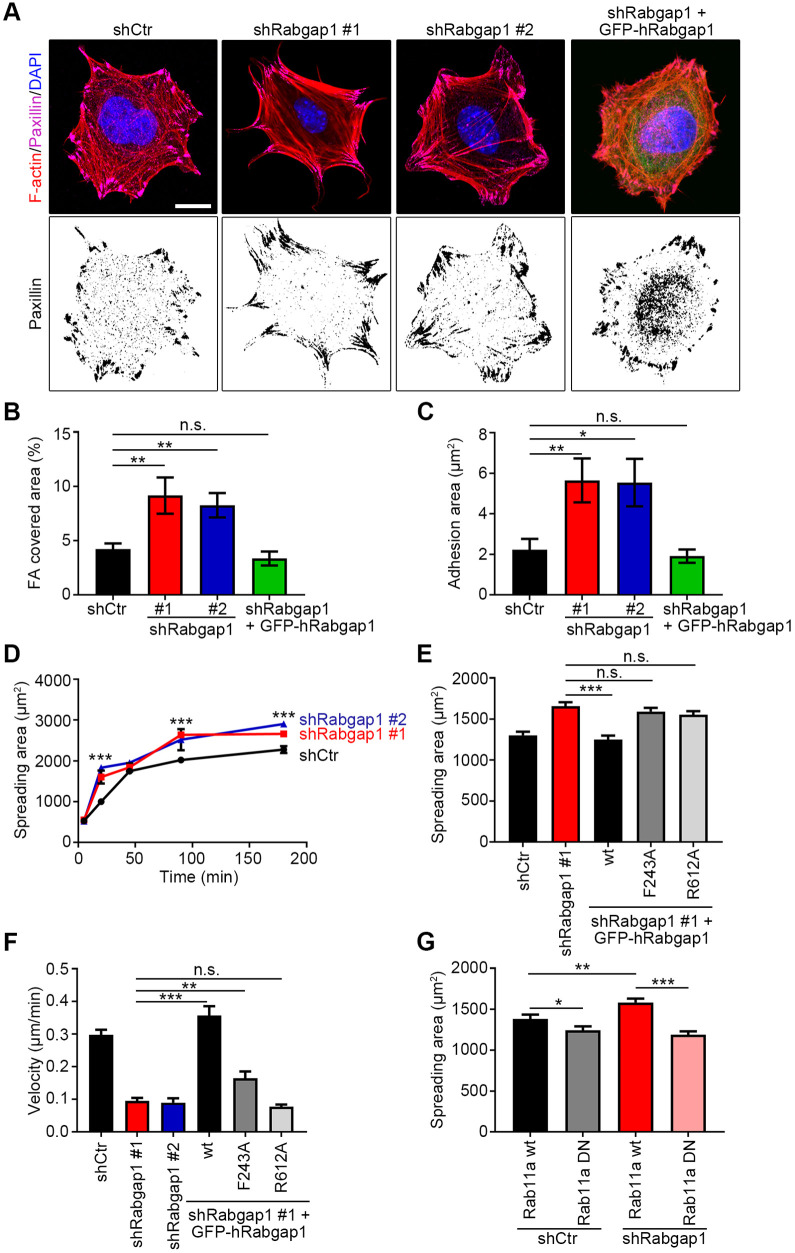
Internalization and intracellular trafficking of β1 integrins are key regulators of cell–ECM interactions and cell motility. To determine whether Rabgap1 regulates β1-integrin-mediated functions, we plated control (shCtr) and Rabgap1-depleted (shRabgap1) mouse fibroblasts on fibronectin and analyzed the number and size of paxillin-positive focal adhesions (Fig. 5A). Loss of Rabgap1 significantly increased the total focal adhesion area and the average focal adhesion size (Fig. 5A–C), and expression of shRNA-resistant GFP-tagged human Rabgap1 in Rabgap1-depleted cells (shRabgap1#1+GFP–hRabgap1) restored these values to control levels (Fig. 5A–C). One explanation for the increased adhesion sizes in Rabgap1-depleted cells, despite reduced levels of active β1 integrin on the cell surface, could be the increased recruitment of β3 integrin into focal adhesions (Fig. S4A,B). Next, we analyzed cell adhesion, spreading and migration in control (shCtr) and Rabgap1-depleted (shRabgap1) fibroblasts. Although cell adhesion to fibronectin was not affected in Rabgap1-depleted cells (Fig. S4C) we observed increased spreading compared to control cells (Fig. 5D,E). Efficient recycling of β1 integrin is essential for cell migration (Böttcher et al., 2012; Caswell et al., 2008; Paul et al., 2015), but how the trafficking of active β1 integrins contributes to cell migration is less understood. Therefore, we monitored the movement of control and Rabgap1-depleted fibroblasts on fibronectin in scratch wound assays and in single-cell migration experiments. Time-lapse video microscopy revealed significantly slower wound closure performance and decreased cell migration speed of Rabgap1-depleted cells (Fig. 5F; Fig. S4D). Importantly, expression of shRNA-resistant GFP-tagged wild-type human Rabgap1 in Rabgap1-depleted cells rescued cell spreading and cell migration to control levels, whereas expression of the integrin-binding-deficient (F243A) or GAP-activity-deficient (R612A) Rabgap1 did not (Fig. 5E,F; Fig. S4D). Finally, we tested whether Rab11 functions downstream of Rabgap1 to regulate integrin-mediated cell spreading. Expression of dominant-negative but not wild-type Rab11a decreased cell spreading in Rabgap1-depleted fibroblasts to control levels (Fig. 5G), suggesting that hyper-activation of Rab11 contributes to the Rabgap1 phenotype. Combined, these data show that Rabgap1-regulated β1 integrin trafficking modulates several integrin-mediated functions in fibroblasts.
Fig. 5.
Rabgap1 regulates β1 integrin-mediated cell spreading and migration. (A) Immunostaining of control (shCtr), Rabgap1-depleted (shRabgap1#1, shRabgap1#2) and GFP-tagged human Rabgap1-expressing rescue (shRabgap1+GFP–hRabgap1) mouse fibroblasts plated on fibronectin for 90 min, using antibodies against paxillin to visualize cell–matrix adhesions. The merge images show the overlay of paxillin (magenta) and F-actin (phalloidin-TRITC, red). DAPI (blue) was used to stain nuclei. Scale bar: 10 μm. (B,C) Rabgap1 depletion increases the percentage of focal adhesion (FA) covered area (B) and the adhesion size (C) of fibroblasts seeded on fibronectin for 90 min. Data are mean±s.e.m. of n=30 cells. *P<0.05; **P<0.01; n.s., not significant (unpaired t-test). (D) Quantification of spreading areas from time-lapse movies of control (shCtr) and Rabgap1-depleted (shRabgap1) fibroblasts seeded on fibronectin for the indicated time periods. Data are mean±s.e.m. of n=10 cells. ***P <0.001 (unpaired t-test). (E) Quantification of spreading areas of the indicated cell lines seeded for 90 min on fibronectin. Data are mean+s.e.m. of n=60 cells. ***P<0.001; n.s., not significant (unpaired t-test). (F) Quantification of single-cell migration velocity of control (shCtr), Rabgap1-depleted (shRabgap1#1 and shRabgap1#2), and GFP–hRabgap1-expressing rescue fibroblasts extracted from time-lapse microscopy recordings by single-cell tracking. Data are mean+s.e.m. of n=28 cells. **P<0.01; ***P <0.001; n.s., not significant (unpaired t-test). (G) Quantification of spreading areas of control (shCtr) or Rabgap1-depleted fibroblasts (shRabgap1) expressing either the GFP-tagged wild-type (Rab11a wt) or dominant-negative Rab11a (Rab11a DN), seeded for 90 min on fibronectin. Data are mean+s.e.m. of n=60 cells. *P<0.05; **P<0.01; ***P<0.001 (unpaired t-test).
Rabgap1 is required for invasive migration in breast cancer cells
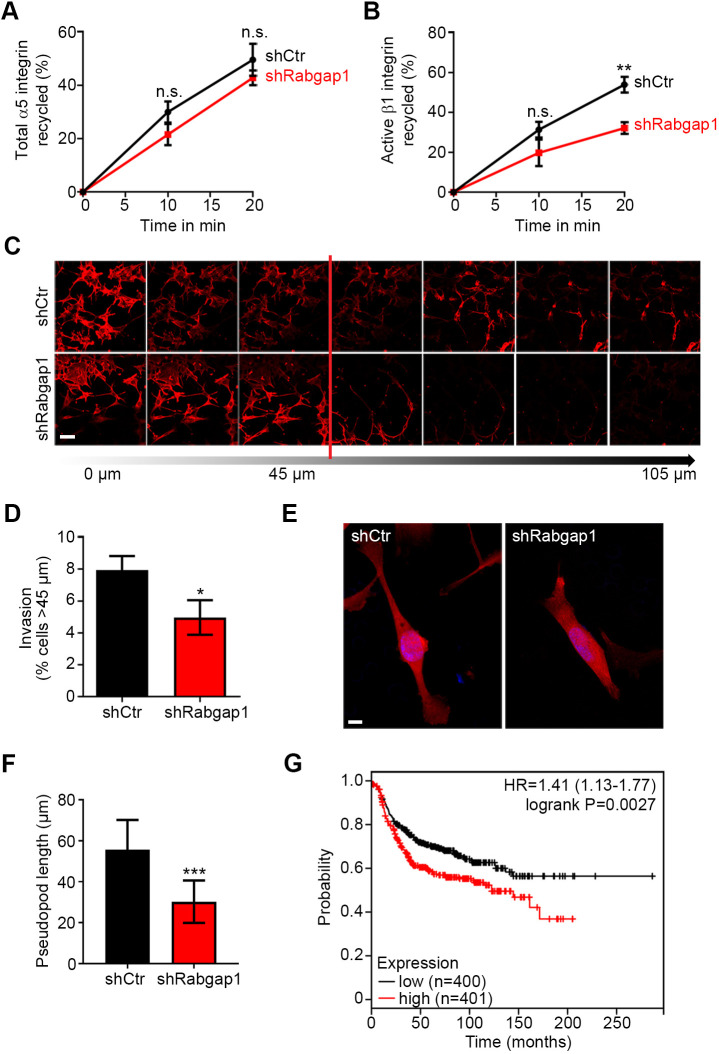
Integrin trafficking does not only regulate cell migration but also cancer cell invasion (Caswell et al., 2008; Paul et al., 2015). Therefore, we tested whether the invasive properties of MDA-MB-231 breast cancer cells depend on Rabgap1 expression. Similar to fibroblasts, depletion of Rabgap1 by short hairpin RNA (shRabgap1) in MDA-MB-231 cells (Fig. S2B) reduced the recycling of active but not total α5β1 integrin to the plasma membrane (Fig. 6A,B). Control MDA-MB-231 cells (shCtr) efficiently migrated through fibronectin-coated membranes and invaded 3D-Matrigel, and this migration and invasion was significantly diminished upon shRNA-mediated depletion of Rabgap1 (Fig. 6C,D). Confocal microscopy analysis of the invading cancer cells revealed that Rabgap1-depleted cells were unable to extend long invasive pseudopods (Fig. 6E,F), formation of which is characteristic of α5β1-integrin-driven invasive migration (Caswell et al., 2008; Jacquemet et al., 2013). These data demonstrate that Rabgap1 promotes invasive migration of a human breast cancer cell line and suggest that high levels of Rabgap1 could represent a prognostic marker for breast cancer patients. To evaluate this possibility, we consulted microarray databases of breast cancer samples (http://kmplot.com/analysis/) (Györffy et al., 2010) and correlated Rabgap1 transcript levels with patient survival. The analyses show that for individuals with estrogen receptor (ER)-negative breast cancer, high expression of Rabgap1 leads to a marked reduction in disease-free survival rate compared to the survival rate of individuals with low Rabgap1 levels (Fig. 6G). Taken together, these results show that Rabgap1 promotes cancer cell invasion in vitro and its elevated mRNA levels are associated with a poor prognosis in human breast cancer patients.
Fig. 6.
Rabgap1 enhances invasive migration in vitro and high Rabgap1 expression correlates with poor prognosis of breast cancer patients. (A,B) Quantification of total (A) and active (B) α5β1 integrin recycling in control (shCtr) or Rabgap1-depleted MDA-MB-231 cells (shRabgap1) by capture ELISA. Data are mean±s.e.m. n=4. **P<0.01; n.s., not significant (unpaired t-test). (C) Confocal microscopy analysis of control (shCtr) and Rabgap1-depleted (shRabgap1) MDA-MB-231 cells migrating into a fibronectin-supplemented Matrigel 3D matrix in an inverted invasion assay. Cells were stained for F-actin using phalloidin–TRITC, and nuclei were labeled with DAPI. Serial optical sections were captured at 15 µm intervals and are presented as a sequence in which the individual optical sections are placed alongside one another, with increasing depth from left to right. Invasion is expressed as the proportion of cells that migrate further than 45 μm (red line). Scale bar: 20 µm. (D) Quantification of the invasiveness of control (shCtr) and Rabgap1-depleted (shRabgap1) MDA-MB-231 cells using the area calculator plugin in ImageJ. The fluorescence intensity of cells invading more than 45 μm was measured and displayed as a percentage of the fluorescence intensity of all cells within the plug. Data are mean±s.d.; n=15 (shCtr) and n=12 (shRabgap1). *P<0.05 (unpaired t-test). (E) Representative confocal images of invading control and Rabgap1-depleted MDA-MB-231 cells stained with phalloidin–TRITC and DAPI to label F-actin and nuclei, respectively. Scale bar: 10 µm. (F) Quantification of pseudopod length of invading control and Rabgap1-depleted MDA-MB-231 cells from confocal images. Data are mean±s.d. n=38. ***P<0.001 (unpaired t-test). (G) Kaplan–Meier analysis indicates a decreased disease-free survival of breast cancer patients with high Rabgap1 expression levels. The analyses show that individuals with high Rabgap1 expression display a significant increase in the hazard ratio (HR) and a pronounced increase in the disease risk compared with individuals with low Rabgap1 expression levels.
DISCUSSION
During their lifetime, α5β1 integrins undergo repeated cycles of endocytosis and exocytosis to dynamically regulate cell adhesion, signaling and motility during homeostasis and in malignant processes such as cancer cell invasion (Caswell et al., 2009; De Franceschi et al., 2015). An emerging concept is that the conformational state of α5β1 integrins affects their trafficking routes. Yet, the underlying molecular mechanisms regulating conformation-specific integrin trafficking remain poorly understood. In this study, we report the characterization of a novel pathway required for the conformation-specific trafficking of β1 integrins. Specifically, we demonstrate that Rabgap1 (1) interacts with the membrane-proximal NPxY motif of the β1 integrin cytoplasmic domain, (2) is required for the efficient recycling of endocytosed active β1 integrin to the plasma membrane, and (3) promotes β1-integrin-mediated migration and invasion of human breast cancer cells. Taken together, our results establish Rabgap1 as a regulatory element in the intracellular trafficking machinery required for α5β1 integrin recycling through its ability to regulate the activity of Rab GTPases, in particular Rab11.
Integrins rely on the binding of proteins to their cytosolic domains to regulate their trafficking through the endosomal system (Böttcher et al., 2012; Caswell et al., 2008; Mana et al., 2016; Pellinen et al., 2006; Steinberg et al., 2012; Tseng et al., 2014). Our proteomics screen for novel cytosolic β1 integrin interactors to the membrane proximal NPxY motif identified not only known interactors such as talins, but also Rabgap1. Subsequent analyses revealed that the interaction of Rabgap1 with β1 integrin depended on the PTB domain in Rabgap1 and the membrane-proximal NPxY motif in the β1 integrin tail. In contrast to a previous study that analyzed a panel of PTB domain-containing proteins for their ability to interact with short integrin β tails in in vitro binding assays (Calderwood et al., 2003), we did not detect an interaction between Rabgap1 and wild-type β3 integrin tails. Functionally, our study showed that Rabgap1 is required for the efficient recycling of endocytosed active β1 integrin to the plasma membrane. This seems surprising, because Rabgap1 was initially identified as a Rab6 GAP associated with centrosomes (Cuif et al., 1999). However, the biological role and subcellular localization of Rabgap1 has not been studied extensively in fibroblasts or cancer cells, and in subsequent biochemical studies Rabgap1 has been shown to regulate Rab4 and Rab11 GTPase activity (Cuif et al., 1999; Fuchs et al., 2007; Kanno et al., 2010), both central players in α5β1 integrin recycling pathways (Caswell et al., 2009). In mouse fibroblasts, we found that a subpopulation of Rabgap1 colocalized with internalized α5β1 integrin in early and recycling endosomes, and we linked aberrant α5β1 integrin trafficking in Rabgap1-depleted cells with irregular Rab11 activation.
Rab GTPases are the master regulators of intracellular membrane traffic and are involved in virtually all steps of trafficking, including vesicle formation, transport and fusion. In addition to Rab4 and Rab11, Rab6 (Shafaq-Zadah et al., 2016), Rab13 (Sahgal et al., 2019), Rab22 (Qu et al., 2016) as well as Rab21 and Rab25 (Dozynkiewicz et al., 2012; Mai et al., 2011; Pellinen et al., 2006) play important roles in distinct integrin trafficking pathways. Rab11a localizes to the endocytic recycling compartment and has been implicated in the control of trafficking of internalized receptors through and from the recycling compartment to the plasma membrane (Campa and Hirsch, 2017). We have shown that Rabgap1 required its GAP activity to promote β1 integrin recycling and cell migration, and that Rabgap1-depleted fibroblasts exhibited increased GTP-bound Rab11 levels. While we cannot rule out that other Rabgap1-regulated GTPases function in Rabgap1-mediated recycling of active β1 integrins, the rescue of intracellular active β1 integrin accumulation and cell spreading observed upon expression of a dominant-negative variant of Rab11a in Rabgap1-depleted cells suggests that hyper-activation of Rab11a contributes to the observed recycling phenotype. Hyper-activation of Rab11 by constitutive activation affects lipid and transferrin trafficking (Ren et al., 1998; Takahashi et al., 2007), and expression of dominant-negative or constitutively-active Rab11 proteins causes abnormal integrin accumulation throughout the cytoplasm in the Drosophila wing as a result of impaired protein trafficking (Bhuin and Roy, 2011). Interestingly, the Rabgap1 paralog Rabgap1L also regulates α5β1 integrin recycling but through inactivation of Rab22a (Qu et al., 2016). However, instead of directly binding to integrin cytosolic domains, Rabgap1L is recruited to phosphatidylinositol (3,4,5)-trisphosphate (PIP3)-positive organelles by AnkB (also known as ANK2), where it inactivates Rab22a to allow α5β1 integrin recycling to the leading edge of migrating fibroblasts (Qu et al., 2016).
We found that Rabgap1 is specifically involved in the regulation of active α5β1 integrin recycling. Although there are still open questions about the conformation of trafficking integrins, several studies have shown, using conformation-specific antibodies, that α5β1 integrins are transported into and through the endo-lysosomal system in inactive (closed) as well as active (extended) conformations. Integrins in different conformations are internalized via distinct mechanisms (Chao and Kunz, 2009; Valdembri et al., 2009) and subsequently take different routes through the endosomal system. While inactive β1 integrin traffics through a Rab4- and actin-dependent short-loop recycling pathway (Roberts et al., 2001) or undergoes retrograde transport to the Golgi apparatus (Shafaq-Zadah et al., 2016), active α5β1 integrin traffics predominantly along the Rab11-dependent long-loop pathway (Arjonen et al., 2012) or is routed for degradation in the absence of CLIC3 (Dozynkiewicz et al., 2012; Kharitidi et al., 2015; Lobert et al., 2010). Our study confirms the involvement of the Rab11-dependent long-loop pathway in the recycling of active α5β1 integrin and could provide a mechanistic explanation underlying the decision between Rab11-dependent recycling and ESCRT-mediated degradation of active α5β1 integrins. Rabgap1 has to associate with the β1 integrin cytosolic domain to efficiently recycle active β1 integrins from endosomal compartments, but how Rabgap1 discriminates between the inactive and the active form of β1 integrin is unclear. Still, it is intriguing to speculate that Rabgap1 binding participates in the decision whether active integrins are recycled in a Rab11-dependent manner or routed into a degradative pathway.
The importance of Rabgap1 for cellular fitness is further emphasized by its ability to regulate cell spreading and to promote cell migration of mouse fibroblasts and cancer cell invasiveness. This requires a functional PTB domain and GAP activity and closely correlates with the ability of Rabgap1 to promote the recycling of active β1 integrins. Still, other Rabgap1-regulated processes and factors, such as β3 integrin redistribution on the cell surface, might contribute to the cellular effects on cell migration and invasion.
In summary, our study describes a role for a complex comprising β1 integrin and Rabgap1 in the recycling of active β1 integrins through the regulation of Rab11 activity and contributes to the remarkable sophistication and specificity of membrane trafficking pathways that determine cargo trafficking in general and specifically integrin transport.
MATERIALS AND METHODS
Antibodies
The following antibodies were used for western blotting (WB) and immunofluorescence (IF): anti-actin (A2066, Sigma-Aldrich; 1:2000 for WB), anti-α5-integrin (555651, Pharmingen; 5 µg/ml ELISA), anti-β1-integrin (MAB1997, MB1.2, Chemicon; 1:400 for IF, 5 µg/ml ELISA), anti-β1-integrin CD29 9EG7 (550531, Pharmingen; 1:100 for IF, 5 µg/ml ELISA), home-made anti-β1-integrin (1:10,000 for WB, 1:2000 for IF; Azimifar et al., 2012), anti-EEA1 (C45B10, Cell Signaling Technology; 1:100 for IF), anti-EGF receptor (#2232, Cell Signaling Technology; 1:1000 for WB), anti-phospho-EGF receptor (Y1068) (#2234, Cell Signaling Technology; 1:1000 for WB), anti-GFP (A11122, Invitrogen; 1:2000 for WB; 11814460001, clones 7.1 and 13.1, Sigma-Aldrich; 1:1000 for WB), anti-kindlin-2 (K3269, Sigma-Aldrich; 1:1000 for WB; MAB2617, clone 3A3, Millipore; 1:1000 for WB), anti-paxillin (610051, clone 349, BD Biosciences; 1:300 for IF), anti-talin (T3287, 8d4, Sigma-Aldrich; 1:1000 for WB), anti-tubulin (MAB 1864, Millipore; 1:1000 for WB), anti-Rabgap1 (ab153992, Abcam; 1:2500 for WB), anti-Rab11 (610656, clone 47, BD Biosciences; 1:1000 for WB), anti-Rab5 (610281, clone 15, BD Biosciences; 1:1000 for WB). Phalloidin–TRITC was used to stain F-actin (P1951, Sigma; 1:600 for IF). The following secondary antibodies were used: goat anti-rabbit Alexa 488-conjugated (A11008), goat anti-mouse Alexa 488-conjugated (A11029), goat anti-rat Alexa 488-conjugated (A11006), goat anti-mouse Alexa 546-conjugated (A11003), goat anti-rabbit Alexa 546-conjugated (A11010) goat anti-rabbit Alexa 647-conjugated (A21244) (1:500 for IF; all from Invitrogen), goat anti-mouse HRP-conjugated (172-1011) and goat anti-rabbit HRP-conjugated (172-1019) (1:10,000 for WB; both from BioRad). DAPI (Sigma) was used to stain nuclei.
Plasmids and constructs
To obtain a GFP-tagged human Rabgap1, the Rabgap1 cDNA (Homo sapiens Rabgap1 mRNA NM_012197.3) was ligated in frame with the pEGFP vector (Clontech) using EcoRI and KpnI restriction enzymes. The Rabgap1–mCherry construct was obtained by exchanging the EGFP sequence for mCherry. For stable depletion of Rabgap1 expression, shRNA target sequences were introduced into the pSuper Retro vector (OligoEngine) to produce retroviral particles: 5′-GAAGAAACTCCTAAAGATA-3′ (shRabgap1#1), 5′-ACTCAAGATTGTAGGAAAT-3′ (shRabgap1#2). Four silent point mutations were introduced by PCR into the human Rabgap1–eGFP sequence recognized by shRabgap1 (5′-GAGGAAACACCTAAGGACA-3′) and the obtained cDNA was subcloned into the pRetroQ-AcGFP-C1 vector (Clontech). Forward and reverse primers containing the silent point mutations were: forward, 5′-AGCTGGTCATAAACAGGACTTTGTCCTTAGGTGTTTCCTCACCCACAACTTGAAAAATGTGGG-3′; and reverse, 5′-CCCACATTTTCAAGTTGTAAATGAGGAAACACCTAAGGACAAAGTCCTGTTTATGACCACAGCT-3′.
Deletion mutants of Rabgap1 (PTB, TBC, CC domains) were prepared via conventional PCR techniques using the following primer sets: NtermFW, 5′-TAAGCACTCGAGCTATGGATGACAAGGCTTCTG-3′; NtermRV, 5′-CCTGCCACAGTCTGGATGAGAATTCTAAGCA-3′; TBCFW, 5′-TAAGCACTCGAGCTATGTCGCAAAGTTCAGTGA-3′; TBCRV, 5′-TGCTTAGAATTCTCAACTAATCTTCATGTTGC-3′; CCFW, 5′-TAAGCACTCGAGCTATGCAGAAGAAGTTGAAAAAATACGA-3′; CCRV, 5′-TGCTTAGAATTCTCAGCAAGTCTCTTTCCCT-3′. PCR products were subcloned into a modified pRetroQ-AcGFP-C1 retroviral vector. The R612A, F243A and F217A point mutations in the human Rabgap1 cDNA were introduced by site-directed mutagenesis using the following primers (the mutations introduced are underlined): R612A fwd, 5′-GCTATCACCCGGGATATTAACGCAACATTCCCAGCCCATGACTAC-3′; R612A rev, 5′-GTAGTCATGGGCTGGGAATGTTGCGTTAATATCCCGGGTGATAGC-3′; F243A fwd, 5′-GTCATTACAATGCAGAGCTCGCCAGAATACACGTCTTCCGGTGT-3′; F243A rev, 5′-ACACCGGAAGACGTGTATTCTGGCGAGCTCTGCATTGTAATGAC-3′; F217A fwd, 5′-TACCCTATCTACAAAATCCTCGCCTGTGTCAGAGGGCATGATGGAACT-3′; F217A rev, 5′-AGTTCCATCATGCCCTCTGACACAGGCGAGGATTTTGTAGATAGGGTA-3′.
The human α5 integrin (ITGA5) cDNA was introduced into the pcDNA3.1-BioID2-HA plasmid obtained from Kyle Roux (University of South Dakota, Vermillion, SD) and the integrin α5–BioID2 sequence subsequently subcloned into the retroviral pLZRS vector (obtained from John G. Collard, The Netherlands Cancer Institute, Amsterdam, The Netherlands). Lamp1–mRFP and Rab7–mRFP plasmids were obtained from Jim Norman (Beatson Institute, Glasgow, UK), the Rab5a–GFP plasmid was provided by Lukas Huber (Medical University Innsbruck, Austria), the Rab11a-S25N–eGFP plasmid was provided by Marino Zerial (MPI-CBG, Dresden, Germany), Rab11a–eGFP and Rab4–eGFP plasmids were obtained from Guido Serini (IRCCS, Candiolo, Italy), Rab11–RFP and Rab4–RFP plasmids were provided by Addgene. Plasmids encoding the C-terminal portion of the Rab11 effector (Rab11-FIP3) cloned in pGex 4T2 vector and the plasmid encoding the Rab5 effector (EEA1 1277-1411) cloned in pGEX-3x vector were kindly provided by Emilio Hirsch (University of Torino, Italy) and Letizia Lanzetti (Institute for Cancer Research and Treatment, Candiolo, Italy), respectively.
Cell lines and transient and stable transfection
Integrin β1 flox/flox mouse fibroblasts and integrin β1 knockout cells re-expressing integrin β1 wt or β1 Y783A were previously described (Böttcher et al., 2012) and were cultured in DMEM (Thermo Fisher Scientific) supplemented with 10% (v/v) FBS at 37°C and 5% CO2. The human breast cancer cell line MDA-MB-231 was purchased from ATCC and cultured in Leibovitz medium (Thermo Fisher Scientific) supplemented with 10% (v/v) FBS at 37°C without CO2. All cell lines were regularly tested for bacterial and mycoplasma contamination.
Cells were transiently transfected using Lipofectamine 2000 (Invitrogen), following the manufacturer's protocol. To generate stable cell lines, VSV-G pseudo-typed retroviral vectors were produced by transient transfection of HEK293T (human embryonic kidney) cells. Viral particles were concentrated from cell culture supernatant, as described previously (Pfeifer et al., 2000), and used for infection.
Integrin tail peptide pulldowns
Pulldowns were performed as described previously (Böttcher et al., 2012) using integrin β1 wt cytoplasmic tail peptides (amino acids 758–798: HDRREFAKFEKEKMNAKWDTGENPIYKSAVTTVVNPKYEGK-OH), integrin β1 Y795A tail peptide (HDRREFAKFEKEKMNAKWDTGENPIYKSAVTTVVNPKAEGK-OH), integrin β1 Y783A tail peptide (HDRREFAKFEKEKMNAKWDTGENPIAKSAVTTVVNPKYEGF-OH), integrin β1 scrambled peptide (EYEFEPDKVDTGAKGTKMAKNEKKFRNYTVHNIWESRKVAP-OH), and integrin β3 wt peptide (HDRKEFAKFEEERARAKWDTANNPLYKEATSTFTNITYRGT). All peptides were desthiobiotinylated at the amino terminus and immobilized on 25 μl Dynabeads MyOne Streptavidin C1 (10 mg ml−1, Invitrogen) before use. Cell lysates were prepared using Mammalian Protein Extraction Reagent (Thermo Fisher Scientific), and 0.5 mg of supernatant was incubated with the peptide-coated beads overnight at 4°C. After several washes of the beads with lysis buffer, the proteins bound to the beads were dissolved in SDS–PAGE Laemmli buffer and analyzed by SDS–PAGE.
Pulldowns from SILAC-labeled cell lysates were performed as described previously (Meves et al., 2011). Briefly, fibroblasts were cultured in the presence of normal medium or in the presence of L-13C615N4-arginine (Arg10)- and L-13C615N2-lysine (Lys8)-labeled medium. 1 mg of unlabeled or Arg10-, Lys8-labeled cell lysate was incubated with β1 wt or β1 Y783A peptide overnight at 4°C. After washing with Mammalian Protein Extraction Reagent (Thermo Fisher Scientific), beads of corresponding peptide pairs were combined, and proteins were eluted by incubating the beads in 16 mM biotin (Sigma-Aldrich) in PBS (pH 7.0) for 30 min at 30°C followed by precipitation with cold acetone at −20°C overnight. The protein pellet was dissolved in SDS–PAGE sample buffer and separated on a 4–15% gradient SDS–PAGE gel. The gel was stained with Coomassie Blue using the Gel Code Blue Safe Protein Stain reagent (Thermo Fisher Scientific) then used for mass analysis. Protein digestion, subsequent peptide fractionation and liquid chromatography-mass spectrometry (LC-MS) analysis were performed as described previously (Meves et al., 2011; Tseng et al., 2014).
BioID assay
Integrin-α5–BioID2-expressing β1 wt and β1 Y783A fibroblasts were treated with 50 µM biotin (Sigma-Aldrich) for 16 h at 37°C. Cells were washed in cold PBS, lysed in β1 IP lysis buffer [50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA and protease inhibitor cocktail (Roche)] and lysates cleared by centrifugation for 15 min at 15,800 g at 4°C. Lysates were incubated with Streptavidin Mag Sepharose beads (GE Healthcare) overnight at 4°C, washed three times with β1 IP lysis buffer and twice with PBS before the isolated biotinylated proteins were analyzed by western blotting.
Pulldown assay for detection of Rab11-GTP and Rab5-GTP
The C-terminal portion of the Rab11 effector FIP3 (Rab11-FIP3) and part of the Rab5 effector EEA1 (amino acids 1277–1411) were produced in bacteria. E. coli Rosetta cells were grown and induced with IPTG (0.5 mM) at 18°C overnight, then lysed, incubated with GST-beads, frozen in liquid nitrogen, and stored at −80°C in 50% glycerol in 50 mM Tris-HCl pH 7.4, 5 mM MgCl2 and 100 mM NaCl.
The pulldown was performed as previously described (Franco et al., 2014). Briefly, the cells were washed twice in ice-cold PBS, lysed in 1 ml of MLB buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% Igepal CA-630, 10% glycerol, 25 mM NaF, 10 mM MgCl2, 1 mM EDTA, 1 mM sodium orthovanadate, and protease inhibitor cocktail) and the lysates cleared by centrifugation for 15 min at 15,800 g at 4°C. A total of 1 mg of cell extract was incubated with 60 μg of recombinant protein coupled to Glutathione Sepharose 4B (GE Healthcare). The reaction mixture was gently rocked for 3 h at 4°C. Beads were washed four times with MLB buffer before samples were resuspended in Laemmli buffer for SDS–PAGE and immunoblot analysis. Endogenous levels of total Rab11 or Rab5 in cell lysates was measured by loading 50 μg of total extracts to normalize measurements of active Rab11 or Rab5. For quantification analysis, ImageJ was used and pictures were taken within the linear intensity range.
FACS analysis
For FACS analysis, fibroblasts were suspended and incubated for 1 h with primary fluorophore-conjugated antibodies on ice and then washed twice with FCS-PBS (3 mM EDTA, 2% FCS in PBS). FACS analysis was carried out using a FACSCanto Cytometer (BD Biosciences) equipped with FACS DiVa software (BD Biosciences). Data analysis was conducted using the FlowJo program (Version 9.4.10).
Immunofluorescence microscopy
For immunostaining, cells were cultured on glass coverslips coated with 10 μg ml−1 fibronectin (Calbiochem). Cells were fixed with 4% PFA in PBS for 15 min at room temperature, washed with PBS and permeabilized with 0.2% Triton X100 in PBS for 10 min at room temperature. Cells were blocked with 3% BSA in PBS for 1 h at room temperature followed by incubation with the primary antibody in PBS containing 3% BSA overnight at 4°C and incubation with secondary antibodies for 1 h at room temperature in the dark. DAPI was used to counterstain nuclei before coverslips were mounted with Elvanol (Mowiol and Dabco, Sigma-Aldrich). Fluorescence images were acquired using an LSM 780 confocal microscope (Zeiss, Germany) equipped with 100×/NA 1.46 oil and 40×/1.4 oil objectives. z-stack projection and contrast adjustments in ImageJ (v1.47) were used for further image analysis.
Integrin trafficking assays using immunofluorescence
To determine the endocytic trafficking of β1 integrins from the cell surface, cells were washed with cold PBS and incubated with a homemade anti-β1-integrin antibody (1:1000) and the 9EG7 antibody (550531, Pharmingen, 1:100) for 30 min on ice. Surface-bound antibody was allowed to internalize for different times at 37°C in regular growth medium. At different time points, the samples were washed with cold PBS and remaining antibodies at the cell surface were removed by two acid washes (0.2 M acetic acid and 0.5 M NaCl in PBS) for 2 min on ice. The serum-induced recycling with 20% FBS was allowed to proceed for different times at 37°C. Subsequently, the cells were fixed with 4% PFA in PBS for 15 min on ice, washed with PBS and permeabilized with 0.01% saponin in PBS for 15 min on ice. Cells were blocked with 3% BSA in PBS for 1 h followed by incubation with the primary antibody in PBS containing 3% BSA and 0.01% saponin overnight at 4°C and incubation with secondary antibodies for 1 h at room temperature in the dark. Nuclei were stained with DAPI before coverslips were mounted with Elvanol. Fluorescence images were acquired with an LSM 780 confocal microscope (Zeiss, Germany) equipped with 100×/NA 1.46 oil and 40×/1.4 oil objectives. For quantification, total fluorescence intensity was scored from the entire cell using the Mean Intensity option in ImageJ, while the percentage of endocytosed β1 integrin was measured from mid-slice confocal images using a region of interest (ROI) drawn inside the cell with the plasma membrane as the boundary. Integrated densities were measured using ImageJ, and the internalized signals were normalized against the total fluorescence signal of the same slice and cell (ROI around the whole cell). Colocalization was analyzed by calculating the Pearson's correlation coefficient using the plugin Coloc2 in Fiji, to restrict the analysis to single cells and to subtract the background signal in a streamlined manner.
Integrin trafficking assays using capture ELISA
Integrin-trafficking assays were performed as described previously with minor modifications (Roberts et al., 2001). Briefly, cells were serum starved for 1 h, transferred to ice, washed twice in cold PBS, and surface labeled at 4°C with 0.2 mg/ml NHS-SS-biotin (Thermo Fisher Scientific) in PBS for 30 min. Labeled cells were washed in cold PBS and transferred to DMEM containing 10% FBS at 37°C to allow internalization. At the indicated times, the medium was aspirated, the dishes were rapidly transferred to ice and washed twice with ice-cold PBS. Biotin was removed from proteins remaining at the cell surface by incubation with a solution containing 20 mM MesNa (Sigma-Aldrich) in 50 mM Tris (pH 8.6) and 100 mM NaCl for 15 min at 4°C. MesNa was quenched by the addition of 20 mM iodoacetamide (Sigma-Aldrich) in PBS for 10 min, and the cells were lysed in lysis buffer (200 mM NaCl, 75 mM Tris-HCl pH 7.5, 15 mM NaF, 1.5 mM Na3VO4, 7.5 mM EDTA, 7.5 mM EGTA, 1.5% Triton X-100, 0.75% Igepal CA-630 and protease inhibitors). Lysates were passed ten times through a 27G needle and clarified by centrifugation at 10,000 g for 10 min. The amount of biotinylated integrins was quantified by capture ELISA.
For recycling assays, internalization was allowed for 30 min at 37°C, and the remaining surface label was stripped using MesNa. Afterwards, cells were incubated in regular growth medium containing 10% FBS for different time points at 37°C, followed by a second stripping using MesNa. Finally, cells were lysed, as above, and subjected to capture ELISA.
Maxisorb 96-well plates (Life Technologies) were coated overnight with 5 μg/ml of anti-β1 integrin antibody (MAB1997, Chemicon) for total mouse β1 integrin, anti-α5 integrin antibody (555651, Pharmingen) for total human α5 integrin or anti-β1 integrin 9EG7 antibody (550531, Pharmingen) for the active β1 integrin in carbonate buffer overnight at 4°C. Nonspecific binding was blocked by incubation with 5% BSA in PBS containing 0.1% Tween-20 (PBS-T) for 1–2 h at room temperature before adding 50 μl cell lysate for incubation overnight at 4°C to capture integrins. Following extensive washes with PBS-T, plates were incubated with streptavidin–HRP (1:1000 in PBS-T containing 1% BSA) for 1 h at 4°C. Biotinylated α5 and β1 integrins were detected by chromogenic reaction with ABTS peroxidase substrate (Vector Laboratories).
Time-lapse video microscopy of cell spreading and random migration
To analyze cell spreading and random migration, 6-well culture dishes were coated with 10 μg ml−1 fibronectin (Calbiochem) and blocked with 1% BSA in PBS. After seeding, phase-contrast video time-lapse microscopy was initiated using a Zeiss Axiovert 200 M (Zeiss, Germany) equipped with 10×/.3 and 20×/.4 objectives, a motorized stage (Märzhäuser) and an environment chamber (EMBL Precision Engineering) with a cooled CCD (charge-coupled device) camera (Roper Scientific).
Spreading cells were imaged in serum-free medium at 5 min intervals for 4 h at 37°C. A total of 20 spreading cells of each investigated cell line from three independent movies were analyzed using ImageJ. Migrating cells were imaged at 10–15 min intervals for 4 h at 37°C. A total of 30 migrating cells of each investigated cell line from three independent movies were analyzed using the manual tracking plugin of ImageJ and the Chemotaxis and Migration Tool (v2.0) of the QWT project. Cell spreading was also measured with cells seeded on fibronectin-coated glass slides, fixed with 4% PFA at 37°C at the indicated time points (20, 45, 90 min) and stained with phalloidin–TRITC to label F-actin.
Scratch wound assay
Scratch wound assays were performed with confluent monolayers of serum-starved cells. 1×105 serum-starved cells were plated in a fibronectin-coated (10 μg ml−1) Ibidi support (Culture-Inserts 80209, Ibidi) in a 6-well dish in serum-free DMEM. Mitomycin C (Sigma-Aldrich, 25 μg/ml) was added for 2 h at 37°C before imaging to limit the proliferation process. Wound closure was imaged in serum-free medium at 15 min intervals overnight at 37°C. Images of live cells were recorded at 37°C and 5% CO2 on a Zeiss Axiovert 200 M (Zeiss, Germany) equipped with 10×/.3 and 20×/.4 objectives, a motorized stage (Märzhäuser) and an environment chamber (EMBL Precision Engineering) with a cooled CCD (charge-coupled device) camera (Roper Scientific). From time-lapse videos, the acquired images of representative wounds were analyzed by calculating percentage of wound closure [defined as the wound area after 12 h relative to the wound area before imaging (0 h)] at the indicated times.
Inverted invasion assay
Inverted invasion assays were carried out as described previously with modifications (Jacquemet et al., 2013). Briefly, the outer surface of cell culture inserts (Transwell; Corning) were coated with fibronectin (final concentration 10 μg/ml in PBS) for 1 h at 37°C. The fibronectin solution was removed and 1×104 MDA-MB-231 cells were plated into the coated inserts followed by a 1.5 h incubation at 37°C. Inserts were inverted and transferred into a 24-well plate containing 1 ml serum-free DMEM per well. Matrigel/matrix solution was prepared on ice by diluting the Matrigel (BND, #354234) with three volumes of DMEM containing 10% FBS and adding fibronectin to a final concentration of 50 μg/ml. Subsequently, the plates were incubated overnight at 37°C, 200 μl of DMEM containing 10% FBS was carefully added to cover the solidified Matrigel matrix. 72 h after seeding, invading cells were fixed and stained with phalloidin–TRITC and DAPI to visualize F-actin and nuclei, respectively. Serial optical sections were captured at 15-μm intervals using an LSM 780 confocal microscope (Zeiss, Germany) and a 40×/1.4 oil objective. Invasion was quantified using ImageJ, measuring the fluorescence intensity of cells invading 45 μm or more, and expressing this as a percentage of the fluorescence intensity of all cells within the plug.
Kaplan–Meier analysis of gene expression microarray
To analyze the prognostic value of Rabgap1 mRNA expression levels, the Kaplan–Meier plotter was used (http://kmplot.com/analysis/) (Györffy et al., 2010). The patient samples (Affy ID: 213313_at; ER negative; PR, HER2, TP53 status: all; n=801) were divided into two groups according to various quantile expressions of Rabgap1 (low and high expression). The two patient groups were compared on the basis of a Kaplan–Meier survival plot, and the hazard ratio with 95% confidence intervals and logrank P-value were calculated.
Statistics
Statistical analyses were performed using GraphPad Prism software (version 5.00, GraphPad Software). Statistical significance was determined using a two-tailed unpaired Student's t-test, comparing the control with each experimental sample. Results are expressed as the mean±s.e.m. unless indicated otherwise.
Supplementary Material
Acknowledgements
We thank Hildegard Reiter for expert technical help. We thank Drs Jim Norman, Emilio Hirsch, Letizia Lanzetti, Kyle Roux, Marino Zerial and Lukas Huber for providing plasmids.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: A.V.S., R.F., R.T.B.; Methodology: A.V.S., A.M., R.T.B.; Validation: A.V.S., R.T.B.; Investigation: A.V.S., T.Z., A.M., R.T.B.; Writing - original draft: A.V.S., R.T.B.; Writing - review & editing: A.V.S., T.Z., A.M., R.F., R.T.B.; Supervision: R.F., R.T.B.; Funding acquisition: R.F., R.T.B.
Funding
The work was supported by the Deutsche Forschungsgemeinschaft (SFB914, project A05 to R.T.B. and R.F.), the National Institutes of Health (K08 CA215105 to A.M.), the Max-Planck-Gesellschaft and the Deutsches Zentrum für Herz-Kreislaufforschung, partner site Munich Heart Alliance. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at https://jcs.biologists.org/lookup/doi/10.1242/jcs.243683.supplemental
Peer review history
The peer review history is available online at https://jcs.biologists.org/lookup/doi/10.1242/jcs.243683.reviewer-comments.pdf
References
- Alanko J., Mai A., Jacquemet G., Schauer K., Kaukonen R., Saari M., Goud B. and Ivaska J. (2015). Integrin endosomal signalling suppresses anoikis. Nat. Cell Biol. 17, 1412-1421. 10.1038/ncb3250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arjonen A., Alanko J., Veltel S. and Ivaska J. (2012). Distinct recycling of active and inactive β1 integrins. Traffic 13, 610-625. 10.1111/j.1600-0854.2012.01327.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimifar S. B., Böttcher R. T., Zanivan S., Grashoff C., Kruger M., Legate K. R., Mann M. and Fässler R. (2012). Induction of membrane circular dorsal ruffles requires co-signalling of integrin-ILK-complex and EGF receptor. J. Cell Sci. 125, 435-448. 10.1242/jcs.091652 [DOI] [PubMed] [Google Scholar]
- Bazzoni G., Shih D.-T., Buck C. A. and Hemler M. E. (1995). Monoclonal antibody 9EG7 defines a novel beta 1 integrin epitope induced by soluble ligand and manganese, but inhibited by calcium. J. Biol. Chem. 270, 25570-25577. 10.1074/jbc.270.43.25570 [DOI] [PubMed] [Google Scholar]
- Bhuin T. and Roy J. K. (2011). Rab11 is required for cell adhesion, maintenance of cell shape and actin-cytoskeleton organization during Drosophila wing development. Int. J. Dev. Biol. 55, 269-279. 10.1387/ijdb.103149tb [DOI] [PubMed] [Google Scholar]
- Böttcher R. T., Stremmel C., Meves A., Meyer H., Widmaier M., Tseng H.-Y. and Fässler R. (2012). Sorting nexin 17 prevents lysosomal degradation of beta1 integrins by binding to the beta1-integrin tail. Nat. Cell Biol. 14, 584-592. 10.1038/ncb2501 [DOI] [PubMed] [Google Scholar]
- Böttcher R. T., Veelders M., Rombaut P., Faix J., Theodosiou M., Stradal T. E., Rottner K., Zent R., Herzog F. and Fässler R. (2017). Kindlin-2 recruits paxillin and Arp2/3 to promote membrane protrusions during initial cell spreading. J. Cell Biol. 216, 3785-3798. 10.1083/jcb.201701176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood D. A., Fujioka Y., de Pereda J. M., Garcia-Alvarez B., Nakamoto T., Margolis B., McGlade C. J., Liddington R. C. and Ginsberg M. H. (2003). Integrin beta cytoplasmic domain interactions with phosphotyrosine-binding domains: a structural prototype for diversity in integrin signaling. Proc. Natl. Acad. Sci. USA 100, 2272-2277. 10.1073/pnas.262791999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood D. A., Campbell I. D. and Critchley D. R. (2013). Talins and kindlins: partners in integrin-mediated adhesion. Nat. Rev. Mol. Cell Biol. 14, 503-517. 10.1038/nrm3624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campa C. C. and Hirsch E. (2017). Rab11 and phosphoinositides: a synergy of signal transducers in the control of vesicular trafficking. Adv. Biol. Regul. 63, 132-139. 10.1016/j.jbior.2016.09.002 [DOI] [PubMed] [Google Scholar]
- Caswell P. T., Chan M., Lindsay A. J., McCaffrey M. W., Boettiger D. and Norman J. C. (2008). Rab-coupling protein coordinates recycling of α5β1 integrin and EGFR1 to promote cell migration in 3D microenvironments. J. Cell Biol. 183, 143-155. 10.1083/jcb.200804140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell P. T., Vadrevu S. and Norman J. C. (2009). Integrins: masters and slaves of endocytic transport. Nat. Rev. Mol. Cell Biol. 10, 843-853. 10.1038/nrm2799 [DOI] [PubMed] [Google Scholar]
- Chao W.-T. and Kunz J. (2009). Focal adhesion disassembly requires clathrin-dependent endocytosis of integrins. FEBS Lett. 583, 1337-1343. 10.1016/j.febslet.2009.03.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Hu J., Yun Y. and Wang T. (2010). Rab36 regulates the spatial distribution of late endosomes and lysosomes through a similar mechanism to Rab34. Mol. Membr. Biol. 27, 23-30. 10.3109/09687680903417470 [DOI] [PubMed] [Google Scholar]
- Cuif M.-H., Possmayer F., Zander H., Bordes N., Jollivet F., Couedel-Courteille A., Janoueix-Lerosey I., Langsley G., Bornens M. and Goud B. (1999). Characterization of GAPCenA, a GTPase activating protein for Rab6, part of which associates with the centrosome. EMBO J. 18, 1772-1782. 10.1093/emboj/18.7.1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Franceschi N., Hamidi H., Alanko J., Sahgal P. and Ivaska J. (2015). Integrin traffic - the update. J. Cell Sci. 128, 839-852. 10.1242/jcs.161653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dozynkiewicz M. A., Jamieson N. B., Macpherson I., Grindlay J., van den Berghe P. V., von Thun A., Morton J. P., Gourley C., Timpson P., Nixon C. et al. (2012). Rab25 and CLIC3 collaborate to promote integrin recycling from late endosomes/lysosomes and drive cancer progression. Dev. Cell 22, 131-145. 10.1016/j.devcel.2011.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eathiraj S., Mishra A., Prekeris R. and Lambright D. G. (2006). Structural basis for Rab11-mediated recruitment of FIP3 to recycling endosomes. J. Mol. Biol. 364, 121-135. 10.1016/j.jmb.2006.08.064 [DOI] [PubMed] [Google Scholar]
- Franco I., Gulluni F., Campa C. C., Costa C., Margaria J. P., Ciraolo E., Martini M., Monteyne D., De Luca E., Germena G. et al. (2014). PI3K class II α controls spatially restricted endosomal PtdIns3P and Rab11 activation to promote primary cilium function. Dev. Cell 28, 647-658. 10.1016/j.devcel.2014.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasa M. A. M., Koessmeier K. T., Ahmadian M. R. and Braga V. M. M. (2012). Illuminating the functional and structural repertoire of human TBC/RABGAPs. Nat. Rev. Mol. Cell Biol. 13, 67-73. 10.1038/nrm3267 [DOI] [PubMed] [Google Scholar]
- Fuchs E., Haas A. K., Spooner R. A., Yoshimura S., Lord J. M. and Barr F. A. (2007). Specific Rab GTPase-activating proteins define the Shiga toxin and epidermal growth factor uptake pathways. J. Cell Biol. 177, 1133-1143. 10.1083/jcb.200612068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromley A., Yeaman C., Rosa J., Redick S., Chen C.-T., Mirabelle S., Guha M., Sillibourne J. and Doxsey S. J. (2005). Centriolin anchoring of exocyst and SNARE complexes at the midbody is required for secretory-vesicle-mediated abscission. Cell 123, 75-87. 10.1016/j.cell.2005.07.027 [DOI] [PubMed] [Google Scholar]
- Györffy B., Lanczky A., Eklund A. C., Denkert C., Budczies J., Li Q. and Szallasi Z. (2010). An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 123, 725-731. 10.1007/s10549-009-0674-9 [DOI] [PubMed] [Google Scholar]
- Hegde S. and Raghavan S. (2013). A skin-depth analysis of integrins: role of the integrin network in health and disease. Cell Commun. Adhes. 20, 155-169. 10.3109/15419061.2013.854334 [DOI] [PubMed] [Google Scholar]
- Hynes R. O. (2002). Integrins: bidirectional, allosteric signaling machines. Cell 110, 673-687. 10.1016/S0092-8674(02)00971-6 [DOI] [PubMed] [Google Scholar]
- Jacquemet G., Morgan M. R., Byron A., Humphries J. D., Choi C. K., Chen C. S., Caswell P. T. and Humphries M. J. (2013). Rac1 is deactivated at integrin activation sites through an IQGAP1-filamin-A-RacGAP1 pathway. J. Cell Sci. 126, 4121-4135. 10.1242/jcs.121988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno E., Ishibashi K., Kobayashi H., Matsui T., Ohbayashi N. and Fukuda M. (2010). Comprehensive screening for novel rab-binding proteins by GST pull-down assay using 60 different mammalian Rabs. Traffic 11, 491-507. 10.1111/j.1600-0854.2010.01038.x [DOI] [PubMed] [Google Scholar]
- Kawasaki N., Isogaya K., Dan S., Yamori T., Takano H., Yao R., Morishita Y., Taguchi L., Morikawa M., Heldin C.-H. et al. (2018). TUFT1 interacts with RABGAP1 and regulates mTORC1 signaling. Cell Discov. 4, 1 10.1038/s41421-017-0001-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharitidi D., Apaja P. M., Manteghi S., Suzuki K., Malitskaya E., Roldan A., Gingras M.-C., Takagi J., Lukacs G. L. and Pause A. (2015). Interplay of endosomal pH and ligand occupancy in integrin alpha5beta1 ubiquitination, endocytic sorting, and cell migration. Cell Rep 13, 599-609. 10.1016/j.celrep.2015.09.024 [DOI] [PubMed] [Google Scholar]
- Kim D. I., Jensen S. C., Noble K. A., Kc B., Roux K. H., Motamedchaboki K. and Roux K. J. (2016). An improved smaller biotin ligase for BioID proximity labeling. Mol. Biol. Cell 27, 1188-1196. 10.1091/mbc.E15-12-0844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. and Springer T. A. (2017). Integrin extension enables ultrasensitive regulation by cytoskeletal force. Proc. Natl. Acad. Sci. USA 114, 4685-4690. 10.1073/pnas.1704171114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobert V. H., Brech A., Pedersen N. M., Wesche J., Oppelt A., Malerød L. and Stenmark H. (2010). Ubiquitination of alpha 5 beta 1 integrin controls fibroblast migration through lysosomal degradation of fibronectin-integrin complexes. Dev. Cell 19, 148-159. 10.1016/j.devcel.2010.06.010 [DOI] [PubMed] [Google Scholar]
- Mai A., Veltel S., Pellinen T., Padzik A., Coffey E., Marjomäki V. and Ivaska J. (2011). Competitive binding of Rab21 and p120RasGAP to integrins regulates receptor traffic and migration. J. Cell Biol. 194, 291-306. 10.1083/jcb.201012126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mana G., Clapero F., Panieri E., Panero V., Böttcher R. T., Tseng H.-Y., Saltarin F., Astanina E., Wolanska K. I., Morgan M. R. et al. (2016). PPFIA1 drives active alpha5beta1 integrin recycling and controls fibronectin fibrillogenesis and vascular morphogenesis. Nat. Commun. 7, 13546 10.1038/ncomms13546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T., Ohbayashi N. and Fukuda M. (2012). The Rab interacting lysosomal protein (RILP) homology domain functions as a novel effector domain for small GTPase Rab36: Rab36 regulates retrograde melanosome transport in melanocytes. J. Biol. Chem. 287, 28619-28631. 10.1074/jbc.M112.370544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meves A., Geiger T., Zanivan S., DiGiovanni J., Mann M. and Fässler R. (2011). Beta1 integrin cytoplasmic tyrosines promote skin tumorigenesis independent of their phosphorylation. Proc. Natl. Acad. Sci. USA 108, 15213-15218. 10.1073/pnas.1105689108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meves A., Stremmel C., Böttcher R. T. and Fässler R. (2013). beta1 integrins with individually disrupted cytoplasmic NPxY motifs are embryonic lethal but partially active in the epidermis. J. Invest. Dermatol. 133, 2722-2731. 10.1038/jid.2013.232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miserey-Lenkei S., Couëdel-Courteille A., Del Nery E., Bardin S., Piel M., Racine V., Sibarita J.-B., Perez F., Bornens M. and Goud B. (2006). A role for the Rab6A’ GTPase in the inactivation of the Mad2-spindle checkpoint. EMBO J. 25, 278-289. 10.1038/sj.emboj.7600929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Layseca P., Icha J., Hamidi H. and Ivaska J. (2019). Integrin trafficking in cells and tissues. Nat. Cell Biol. 21, 122-132. 10.1038/s41556-018-0223-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser M., Legate K. R., Zent R. and Fässler R. (2009). The tail of integrins, talin, and kindlins. Science 324, 895-899. 10.1126/science.1163865 [DOI] [PubMed] [Google Scholar]
- Muller P. A. J., Caswell P. T., Doyle B., Iwanicki M. P., Tan E. H., Karim S., Lukashchuk N., Gillespie D. A., Ludwig R. L., Gosselin P. et al. (2009). Mutant p53 drives invasion by promoting integrin recycling. Cell 139, 1327-1341. 10.1016/j.cell.2009.11.026 [DOI] [PubMed] [Google Scholar]
- Nader G. P. F., Ezratty E. J. and Gundersen G. G. (2016). FAK, talin and PIPKIγ regulate endocytosed integrin activation to polarize focal adhesion assembly. Nat. Cell Biol. 18, 491-503. 10.1038/ncb3333 [DOI] [PubMed] [Google Scholar]
- Pan X., Eathiraj S., Munson M. and Lambright D. G. (2006). TBC-domain GAPs for Rab GTPases accelerate GTP hydrolysis by a dual-finger mechanism. Nature 442, 303-306. 10.1038/nature04847 [DOI] [PubMed] [Google Scholar]
- Paul N. R., Allen J. L., Chapman A., Morlan-Mairal M., Zindy E., Jacquemet G., Fernandez del Ama L., Ferizovic N., Green D. M., Howe J. D. et al. (2015). alpha5beta1 integrin recycling promotes Arp2/3-independent cancer cell invasion via the formin FHOD3. J. Cell Biol. 210, 1013-1031. 10.1083/jcb.201502040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellinen T., Arjonen A., Vuoriluoto K., Kallio K., Fransen J. A. M. and Ivaska J. (2006). Small GTPase Rab21 regulates cell adhesion and controls endosomal traffic of β1-integrins. J. Cell Biol. 173, 767-780. 10.1083/jcb.200509019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer A., Kessler T., Silletti S., Cheresh D. A. and Verma I. M. (2000). Suppression of angiogenesis by lentiviral delivery of PEX, a noncatalytic fragment of matrix metalloproteinase 2. Proc. Natl. Acad. Sci. USA 97, 12227-12232. 10.1073/pnas.220399597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powelka A. M., Sun J., Li J., Gao M., Shaw L. M., Sonnenberg A. and Hsu V. W. (2004). Stimulation-dependent recycling of integrin β1 regulated by ARF6 and Rab11. Traffic 5, 20-36. 10.1111/j.1600-0854.2004.00150.x [DOI] [PubMed] [Google Scholar]
- Pozzi A. and Zent R. (2013). Integrins in kidney disease. J. Am. Soc. Nephrol. 24, 1034-1039. 10.1681/ASN.2013010012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu F., Lorenzo D. N., King S. J., Brooks R., Bear J. E. and Bennett V. (2016). Ankyrin-B is a PI3P effector that promotes polarized alpha5beta1-integrin recycling via recruiting RabGAP1L to early endosomes. Elife 5, e20417 10.7554/eLife.20417.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainero E., Howe J. D., Caswell P. T., Jamieson N. B., Anderson K., Critchley D. R., Machesky L. and Norman J. C. (2015). Ligand-occupied integrin internalization links nutrient signaling to invasive migration. Cell Rep 10, 398-413. 10.1016/j.celrep.2014.12.037 [DOI] [PubMed] [Google Scholar]
- Ren M., Xu G., Zeng J., De Lemos-Chiarandini C., Adesnik M. and Sabatini D. D. (1998). Hydrolysis of GTP on rab11 is required for the direct delivery of transferrin from the pericentriolar recycling compartment to the cell surface but not from sorting endosomes. Proc. Natl. Acad. Sci. USA 95, 6187-6192. 10.1073/pnas.95.11.6187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts M., Barry S., Woods A., van der Sluijs P. and Norman J. (2001). PDGF-regulated rab4-dependent recycling of αvβ3 integrin from early endosomes is necessary for cell adhesion and spreading. Curr. Biol. 11, 1392-1402. 10.1016/S0960-9822(01)00442-0 [DOI] [PubMed] [Google Scholar]
- Sahgal P., Alanko J., Icha J., Paatero I., Hamidi H., Arjonen A., Pietila M., Rokka A. and Ivaska J. (2019). GGA2 and RAB13 promote activity-dependent beta1-integrin recycling. J. Cell Sci. 132, jcs233387 10.1242/jcs.233387 [DOI] [PubMed] [Google Scholar]
- Schiller H. B., Hermann M.-R., Polleux J., Vignaud T., Zanivan S., Friedel C. C., Sun Z., Raducanu A., Gottschalk K.-E., Théry M. et al. (2013). β1- and αv-class integrins cooperate to regulate myosin II during rigidity sensing of fibronectin-based microenvironments. Nat. Cell Biol. 15, 625-636. 10.1038/ncb2747 [DOI] [PubMed] [Google Scholar]
- Shafaq-Zadah M., Gomes-Santos C. S., Bardin S., Maiuri P., Maurin M., Iranzo J., Gautreau A., Lamaze C., Caswell P., Goud B. et al. (2016). Persistent cell migration and adhesion rely on retrograde transport of β1 integrin. Nat. Cell Biol. 18, 54-64. 10.1038/ncb3287 [DOI] [PubMed] [Google Scholar]
- Steinberg F., Heesom K. J., Bass M. D. and Cullen P. J. (2012). SNX17 protects integrins from degradation by sorting between lysosomal and recycling pathways. J. Cell Biol. 197, 219-230. 10.1083/jcb.201111121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y., Xia W., Li J., Walz T., Humphries M. J., Vestweber D., Cabanas C., Lu C. and Springer T. A. (2016). Relating conformation to function in integrin α5β1. Proc. Natl. Acad. Sci. USA 113, E3872-E3881. 10.1073/pnas.1605074113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Tseng H.-Y., Tan S., Senger F., Kurzawa L., Dedden D., Mizuno N., Wasik A. A., Thery M., Dunn A. R. et al. (2016). Kank2 activates talin, reduces force transduction across integrins and induces central adhesion formation. Nat. Cell Biol. 18, 941-953. 10.1038/ncb3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Costell M. and Fässler R. (2019). Integrin activation by talin, kindlin and mechanical forces. Nat. Cell Biol. 21, 25-31. 10.1038/s41556-018-0234-9 [DOI] [PubMed] [Google Scholar]
- Takahashi M., Murate M., Fukuda M., Sato S. B., Ohta A. and Kobayashi T. (2007). Cholesterol controls lipid endocytosis through Rab11. Mol. Biol. Cell 18, 2667-2677. 10.1091/mbc.e06-10-0924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodosiou M., Widmaier M., Böttcher R. T., Rognoni E., Veelders M., Bharadwaj M., Lambacher A., Austen K., Muller D. J., Zent R. et al. (2016). Kindlin-2 cooperates with talin to activate integrins and induces cell spreading by directly binding paxillin. Elife 5, e10130 10.7554/eLife.10130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng H.-Y., Thorausch N., Ziegler T., Meves A., Fässler R. and Böttcher R. T. (2014). Sorting nexin 31 binds multiple β integrin cytoplasmic domains and regulates beta1 integrin surface levels and stability. J. Mol. Biol. 426, 3180-3194. 10.1016/j.jmb.2014.07.003 [DOI] [PubMed] [Google Scholar]
- Tubbesing K., Ward J., Abini-Agbomson R., Malhotra A., Rudkouskaya A., Warren J., Lamar J., Martino N., Adam A. P. and Barroso M. (2020). Complex Rab4-mediated regulation of endosomal size and EGFR activation. Mol. Cancer Res. 18, 757-773. 10.1158/1541-7786.MCR-19-0052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlik M. T., Temple B., Bencharit S., Kimple A. J., Siderovski D. P. and Johnson G. L. (2005). Structural and evolutionary division of phosphotyrosine binding (PTB) domains. J. Mol. Biol. 345, 1-20. 10.1016/j.jmb.2004.10.038 [DOI] [PubMed] [Google Scholar]
- Valdembri D., Caswell P. T., Anderson K. I., Schwarz J. P., König I., Astanina E., Caccavari F., Norman J. C., Humphries M. J., Bussolino F. et al. (2009). Neuropilin-1/GIPC1 signaling regulates α5β1 integrin traffic and function in endothelial cells. PLoS Biol. 7, e25 10.1371/journal.pbio.1000025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegener K. L., Partridge A. W., Han J., Pickford A. R., Liddington R. C., Ginsberg M. H. and Campbell I. D. (2007). Structural basis of integrin activation by talin. Cell 128, 171-182. 10.1016/j.cell.2006.10.048 [DOI] [PubMed] [Google Scholar]
- White D. P., Caswell P. T. and Norman J. C. (2007). alpha v beta3 and alpha5beta1 integrin recycling pathways dictate downstream Rho kinase signaling to regulate persistent cell migration. J. Cell Biol. 177, 515-525. 10.1083/jcb.200609004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winograd-Katz S. E., Fässler R., Geiger B. and Legate K. R. (2014). The integrin adhesome: from genes and proteins to human disease. Nat. Rev. Mol. Cell Biol. 15, 273-288. 10.1038/nrm3769 [DOI] [PubMed] [Google Scholar]
- Ye F., Hu G., Taylor D., Ratnikov B., Bobkov A. A., McLean M. A., Sligar S. G., Taylor K. A. and Ginsberg M. H. (2010). Recreation of the terminal events in physiological integrin activation. J. Cell Biol. 188, 157-173. 10.1083/jcb.200908045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Zhu J. and Springer T. A. (2013). Complete integrin headpiece opening in eight steps. J. Cell Biol. 201, 1053-1068. 10.1083/jcb.201212037 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.