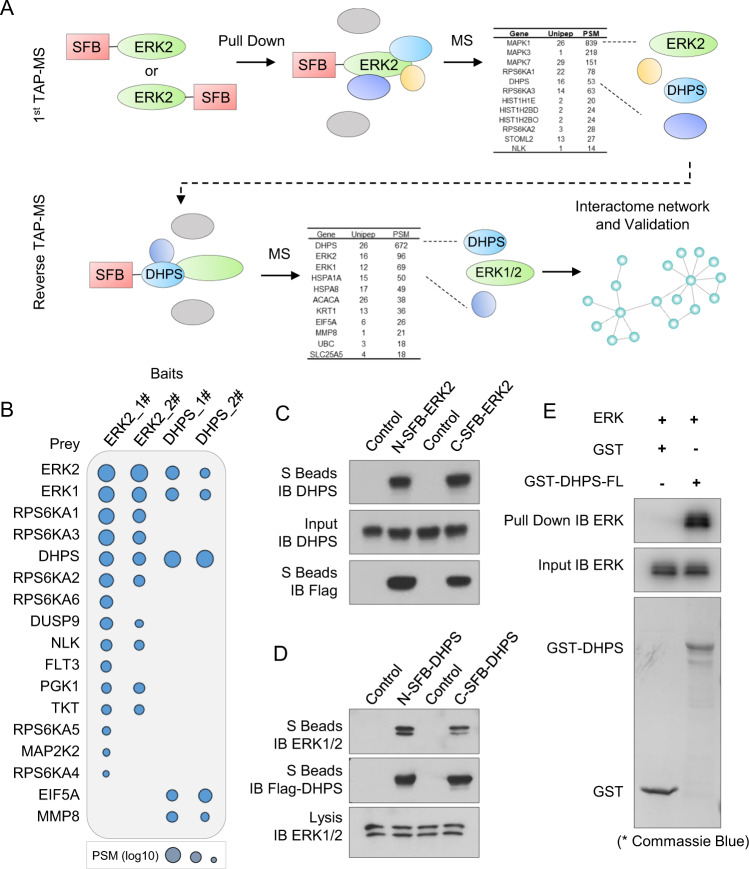
Fig. 1. Reciprocal TAP-MS Analysis Uncovered DHPS and ERK2 Interactome.
a Workflow of the protein interactome study using the TAP-MS approach. Constructs encoding N- or C-terminal SFB-tagged ERK2 were expressed in HEK293T cells. Cell lysates underwent two-step affinity purification using streptavidin beads and S-protein agarose beads. The samples were then analyzed by MS, and the identified peptides were filtered using HCIP analysis. The potential ERK2 interactor DHPS was fused to N- or C-terminal SFB tag and underwent the same TAP-MS analysis. b Visualization of the reciprocal TAP-MS results of ERK and DHPS. The HCIP list for each bait is displayed as a dot plot. The PSMs (log10) of the identified proteins are determined by the size of the circles. c ERK2 interacts with DHPS. N- or C-terminal SFB-tagged ERK2 was expressed in HEK293T cells. Cell lysates were pulled down with S protein beads, and the indicated proteins were detected by Western blotting. Data represent a representative experiment from three independent experiments. d Association between DHPS and ERK1/2. N- or C-terminal SFB-tagged DHPS were expressed in HEK293T cells. Cell lysates were pulled down with S protein beads, and the indicated proteins were detected by Western blotting. Data represent a representative experiment from three independent experiments. e In vitro interaction between DHPS and ERK. ERK protein was incubated with GST protein or GST-DHPS protein. The pull-down assays were performed with glutathione agarose, and the indicated proteins were detected by Western blotting or Coomassie Blue staining.