Abstract
Background
MicroRNAs have been identified as major regulators and therapeutic targets of glioblastoma (GBM). It is thus meaningful to study the miRNAs differentially expressed (DE-miRNAs) in GBM.
Materials and Methods
We performed a meta-analysis of previously published microarray data using the R-based “metaMA” package to identify DE-miRNAs.The biological processes of the DE-miRNAs were then analyzed using FunRich. KEGG pathways of the DE-miRNAs gene targets were analyzed by mirPath V.3. Luciferase activity assay was performed to validate that OXSM is a direct target of hsa-miR338-3p. Flow cytometry was used to detect the effects of miR-338-3p on GBM cell proliferation, apoptosis and cell cycle.
Results
DE-miRNAs in blood and brain tissue from GBM were identified. “Type I interferon signaling pathway” and “VEGF and VEGFR signaling network” were the most significantly enriched biological processes shared by all GBM types. In KEGG pathway analysis, DE-miRNAs both in blood and tissue show altered fatty acid biosynthesis. Further validation shows hsa-miR-338-3p regulates fatty acid metabolism by directly targeting OXSM gene. In addition, our data revealed an accelerated cell cycle and an anti-apoptotic role for OXSM in glioma cells, which has not been reported. Finally, we confirmed that hsa-miR-338-3p inhibitor antagonized the effect of downregulation of OXSM on cell cycle and apoptosis of GBM cells.
Conclusion
We revealed that hsa-miR-338-3p, down-regulated in GBM, may affect the biogenesis and rapid proliferation of glioma cells by regulating the level of OXSM, providing new insights into understanding the pathogenesis of GBM and developing strategies to improve GBM prognosis.
Keywords: glioblastoma, hsa-miR-338-3p, OXSM, fatty acid metabolism
Introduction
Glioblastoma (GBM) is one of the most prevalent and highly invasive malignant tumors.1 The overall survival (OS) time for GBM patients is 15 months and the 5-year survival rate is below 10%, even after comprehensive treatments such as surgery, radiation therapy, and chemotherapy.2,3 This is mainly due to the ability of the tumor in infiltrating the surrounding brain tissues.
MicroRNAs (miRNAs), a class of small non-coding RNAs, play a pivotal role in RNA silencing and post-transcriptional regulation of gene expression, thereby taking part in numerous biological processes and biological pathways. Numerous experiments have found that miRNAs were involved in all aspects of tumor-related processes, including proliferation, apoptosis, metastasis, angiogenesis, and immune response.4–6 Because of its high sensitivity and specificity, miRNA is very likely to become a new type of tumor diagnostic and prognostic markers and a new target for tumor treatment.7,8 Therefore, it is meaningful to study the miRNAs differentially expressed (DE-miRNAs) in GBM including in serum or tumor tissues. In past decades, numerous studies have been conducted to understand the complex host-miRNA interactions of GBM. For example, the expression level of hsa-miR-210-3p in GBM tumor tissue is higher than that in normal brain tissue, and it is positively correlated with the pathological grade of patients9 The hsa-miR-199a-3p inhibits glioma growth by inhibiting AKT/mTOR signaling pathway10 The hsa-miR-96 activates the Wnt/β-catenin pathway by targeting the GSK3b/β-catenin downstream tumor suppressor gene HBP-1 and promoting the proliferation of glioma cells11 The hsa-miR-181b-5p targets IGF-1R and inhibits glioma cell proliferation, migration, and invasion12 The hsa-miR-7-5p might control growth of GBM microvasculature by regulating the PI3K/ATK and Raf/MEM/ERK pathways.13 Although the accumulating data provide useful information about miRNAs in GBM, the identification of key miRNAs and pathways from these studies was restricted due to the limited sample size in the independent study.
Herein, we first screened several DE-miRNAs in blood or tumor tissues from GBM compared with healthy controls or adjacent normal tissues from GBM by analyzing nine independent microarray datasets downloaded from the GEO database. Then, FunRich was employed to predict the biological processes and biological pathways of DE-miRNAs in GBM. Next, KEGG pathway enrichment analysis of genes target by DE-miRNAs in GBM was conducted by mirPath V.3. Subsequently, fatty acid metabolism regulated by DE-miRNAs was further analyzed, GEPIA was used to analyze the mRNA expression related to fatty acid metabolism. In addition, OXSM was identified as a direct target of has-miR-338-3p by luciferase activity analysis. Finally, our data revealed an accelerated cell cycle and an anti-apoptotic role for OXSM in glioma cells, which has not been reported. Considering that hsa-miR-338-3p can down-regulate the expression of the target gene OXSM, we also investigated whether hsa-miR-338-3p is involved in the regulation of GBM cell function by OXSM. We confirmed that hsa-miR-338-3p inhibitor antagonized the effect of downregulation of OXSM on cell cycle and apoptosis of U87 and U251 cells. Our results indicate that hsa-miR-338-3p may affect the biogenesis and rapid proliferation of glioma cells by regulating the level of OXSM.
Our study provides a comprehensive evaluation of miRNA expression profiles, highlights the importance of hsa-miR-338-3p and OXSM in GBM, which will provide new insights into understanding the pathogenesis of GBM and developing strategies to improve GBM prognosis.
Materials and Methods
Microarray Data Collection
The gene expression microarray datasets with the keywords “GBM glioblastoma”, “glioma”, “miRNA” and “microRNA” were downloaded from Gene Expression Omnibus (NCBI) database.14 Glioblastoma patients were considered “Cancer group” while healthy controls or adjacent normal tissues from GBM were considered “Control group”. Data sets with a sample source other than serum, plasma, or brain tissue from the control group were excluded. Nine independent microarray datasets with raw data were selected and the details about these datasets are outlined in Table 1. The following information was extracted from each of the studies that were selected: GEO accession; numbers of patients and controls; sample source; platform and gene expression data. We compared microarray data for GBM (n = 332) and controls (n = 213) from the public database submissions. Three datasets included the transcriptome profiles of serum from 207 GBM and 181 controls. Six datasets included tissue from 125 GBM and 32 controls.
Table 1.
Summary of Transcriptome Datasets Used in This Study
| Study | GEO Accession | Sample Size | Sample Source | Platform | |
|---|---|---|---|---|---|
| Cancer | Control | ||||
| 1 | GSE93850 | 22 | 8 | Serum | GPL22948 State Key Laboratory Human microRNA array 1858 |
| 2 | GSE122488 | 12 | 16 | Serum | GPL11154 Illumina HiSeq 2000 (Homo sapiens) |
| 3 | GSE139031 | 173 | 157 | Serum | GPL21263 3D-Gene Human miRNA V21_1.0.0 |
| 4 | GSE25631 | 82 | 5 | Tissue | GPL8179 Illumina Human v2 MicroRNA expression beadchip |
| 5 | GSE37737 | 7 | 7 | Tissue | GPL9460 Applied Biosystems Human TaqMan Low Density Array |
| 6 | GSE61710 | 12 | 5 | Tissue | GPL10656 Agilent-029297 Human miRNA Microarray v14 Rev.2 |
| 7 | GSE65626 | 3 | 3 | Tissue | GPL19117 [miRNA-4] Affymetrix Multispecies miRNA-4 Array |
| 8 | GSE90603 | 16 | 7 | Tissue | GPL21572 [miRNA-4] Affymetrix Multispecies miRNA-4 Array |
| 9 | GSE103229 | 5 | 5 | Tissue | GPL18058 Exiqon miRCURY LNA microRNA array, 7th generation |
miRNA Expression Analysis
To identify DE-miRNAs between GBM and controls, the data collected from each eligible microarray study were imported to the R-based “metaMA” package developed by Guillemette Marot15 which combines either p-values or modified effect sizes from different studies to find differentially expressed genes. The data were annotated after converting the probe IDs to the corresponding miRNA names. The intensity values for each probe set were log2 transformed then uploaded, processed, and annotated for data integrity. A p-value <0.05 was considered statistically significant for the analysis.
Biological Process and Pathway Enrichment
Gene Ontology classification of miRNAs expressed differentially in GBM was performed with FunRich (http://www.funrich.org/), a stand-alone software tool used mainly for functional enrichment and interaction network analysis based on default background database or a customized database. miRNA id lists were uploaded and analyzed using the functional annotation chart for biological processes and pathways. Representative biological processes and pathways selected from the top significantly enriched charts are reported in the figures. A Benjamini-corrected p-value less than 0.05 was used to identify a statistically significant analysis.
KEGG pathways of genes target by DE-miRNAs in GBMs were analyzed by mirPath V.3(http://diana.imis.athena-innovation.gr/), a stand-alone software tool that can utilize predicted miRNA targets (in the CDS or 3ʹ-UTR region) provided by the microT-CDS algorithm and miRNA interactions experimentally verified derived from TarBase.
Cell Culture and Transfection
The human GBM cell lines U87 and U251 were purchased from the Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, Shanghai, China. The cells were incubated at 37°C, 5% CO2, and 10% fetal bovine serum (Invitrogen) was added to the culture medium. Transfection reagent Lipofectamine 2000 (Invitrogen) was used to transfect 20µM has-miR338-3p mimics or controls (GenePharma, Shanghai, China) into GBM cells according to the manufacturer’s instructions.
Reverse Transcription and qRT-PCR
MiRNeasy Micro kit (QIAGEN) was used to extract miRNA from cells, and purified RNA was treated with DNase I reagent to eliminate genomic DNA contamination. RNA was reverse transcribed using Primpscript® RT kit (TAKARA, Dalian, China) according to the manufacturer’s instructions. qRT-PCR was used to detect miRNA and mRNA using SYBR Premix Ex Taq ™ II (TAKARA, Dalian, China). The miRNA expression level was normalized to small nucleolar RNA (U6), and mRNA expression was normalized to GAPDH.
Luciferase Activity Assay
According to the target prediction, the OXSM 3′UTR contained two appropriate sequences with has-miR338-3p binding sites (358–368 and 437–445 nt). A 3ʹUTR fragment containing the putative binding site of hsa-miR-338-3p was cloned into the GP-miRGLO vector. Lipofectamine 2000 (Invitrogen) was used to transfected empty vector plasmid (GP-miRGLO) or OXSM 3ʹUTR reporter gene (wild type, MUT-1, MUT-2), into U87 and U251 cells. Drosophila and Renilla luciferase activity was measured using the Dual-Luciferase reporter assay (Promega) at 24 h after transfection.
Cell Cycle Assay
Cell cycle phases were determined using the BD Cycletest™ Plus DNA Reagent Kit (BD Biosciences) according to the instructions provided by the manufacturer. In brief, GBM cells were cultured for 48 h after transfection. After transfection, the distribution of DNA content was determined using a BD LSR II flow cytometer and analyzed using the FlowJo software.
Detection of Apoptosis
After transfection (48 h), GBM cells were stained with PE-conjugated anti-Annexin V and 7-AAD (Biolegend). The cells were analyzed using a BD LSR II flow cytometer and the FlowJo software.
Statistical Analysis
Data analysis was performed using SPSS 21.0 and GraphPad Prism Version 5.0 software. A p-value <0.05 was considered statistically significant.
Result
MiRNAs Expressed Differentially in Blood and Brain Tissue from Controls
First, we identified the miRNAs expressed differentially in blood from controls. According to the results of our analysis, 390 miRNAs in blood were identified to be expressed differentially between GBM and controls. Among the 390 miRNAs, 190 miRNAs were upregulated and 200 were downregulated. The downregulated miRNAs included circulating miRNAs that have been reported to be associated with GBM disease progression or pathogenesis, such as hsa-miR-128.16 There were 317 DE-miRNAs in brain tissues between GBM and controls across microarray datasets. Among the 317 miRNAs, 195 miRNAs were upregulated and 122 miRNAs were downregulated. The upregulated miRNAs include hsa-miR-155-5p, which can promote proliferation, invasion, migration, and inhibit apoptosis of glioma cells.17–19 The miRNAs expressed differentially in blood and tissue are shown in Supplemental Table 1.
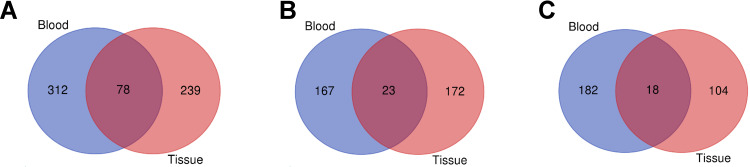
Next, we investigated whether some different miRNAs were shared when comparing different types of samples between GBM and controls. The Venn diagram shows that 78 miRNAs have changed significantly in blood and tissue samples. Among them, 23 up-regulated miRNAs and 18 down-regulated miRNAs showed the same variation trend (Figure 1). Twenty-three upregulated miRNAs and the 18 downregulated miRNAs shared in all sample types are shown in Table 2.
Figure 1.
Venn diagram representing the number of DE-miRNAs in GBM compared with controls in blood and tissue. (A) All the DE-miRNAs found in blood and tissues; (B) the upregulated miRNAs found in blood and tissues; (C) the downregulated miRNAs found in blood and tissue.
Table 2.
miRNAs Expressed Differentially in All Sample Types
| UP | hsa-miR-517c-3p | hsa-miR-4429 | hsa-miR-320a |
| hsa-miR-20b-3p | hsa-miR-3646 | hsa-miR-301b | |
| hsa-miR-2114-5p | hsa-miR-4477a | hsa-miR-891a | |
| hsa-miR-3667-5p | hsa-miR-15b-5p | hsa-miR-188-5p | |
| hsa-miR-675-3p | hsa-miR-4528 | hsa-miR-20b-5p | |
| hsa-miR-195-5p | hsa-miR-19a-3p | hsa-miR-106b-5p | |
| hsa-miR-10b-5p | hsa-miR-4753-3p | hsa-miR-21-3p | |
| hsa-miR-4763-5p | hsa-miR-19b-3p | ||
| DOWN | hsa-miR-323b-5p | hsa-miR-330-3p | hsa-miR-338-5p |
| hsa-miR-138-1-3p | hsa-miR-138-5p | hsa-miR-485-3p | |
| hsa-miR-425-3p | hsa-miR-129-5p | hsa-miR-338-3p | |
| hsa-miR-490-3p | hsa-miR-4270 | hsa-miR-5589-5p | |
| hsa-miR-330-5p | hsa-miR-2116-5p | hsa-miR-5187-5p | |
| hsa-miR-139-5p | hsa-miR-1225-5p | hsa-miR-138-2-3p |
MiRNAs Functional Classification and Pathway Assignment
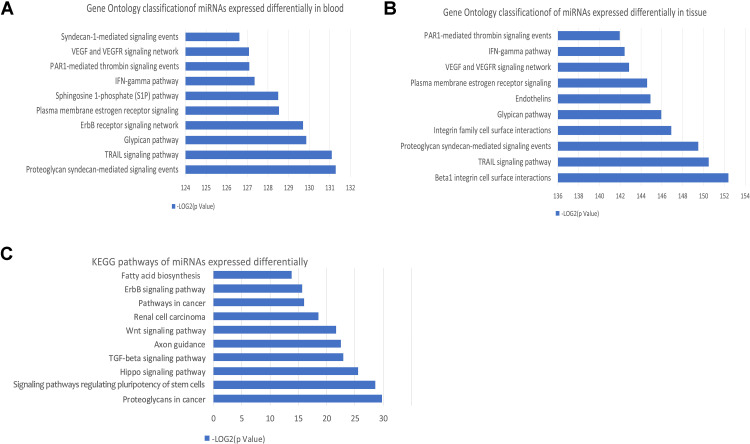
Gene Ontology classification analysis was carried out for 390 DE-miRNAs in blood or 317 DE-miRNAs in brain tissues. Following GO analysis, the “TRAIL signaling pathway”, “VEGF and VEGFR signaling network” and “IFN-gamma pathway” were significantly enriched for miRNAs expressed differentially in blood and tissue (Figure 2A and B). KEGG’s PATHWAY database integrates current knowledge in molecular interaction networks. In this study, we analyzed KEGG pathways of 78 DE-miRNAs both in blood and tissue (Figure 2C), revealing that “Fatty acid biosynthesis” was significantly enriched.
Figure 2.
GO and KEGG pathway analysis of DE-miRNAs in GBM. (A) GO classification of genes targeted by 390 DE-miRNAs in blood. (B) GO classification of genes targeted by 317 DE-miRNAs in tissue. (C) KEGG pathways of 78 DE-miRNAs in all type of samples.
Fatty Acid Metabolism Regulated by miRNAs
It has become increasingly clear that patients with high-risk/relapsed tumors are intimately linked to metabolic abnormalities, as well as abnormal expression of multiple factors and signaling pathways related to tumor metabolism. Fatty acid biosynthesis has been reported to play important roles in the pathogenesis of multi cancers.20
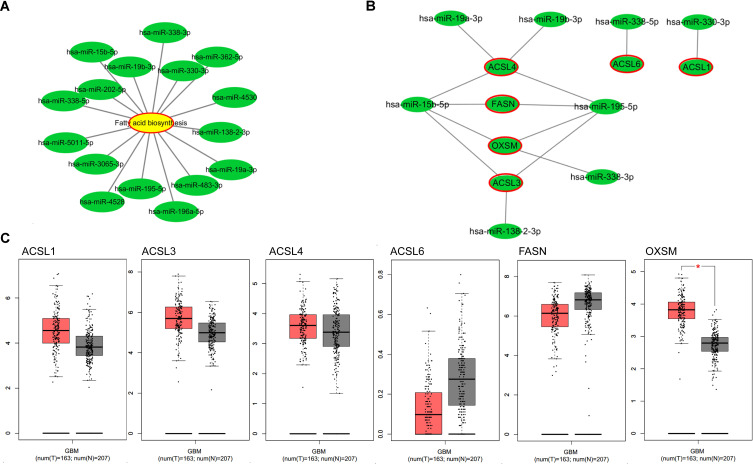
In the KEGG pathway analysis, DE-miRNAs in both blood and tissues may affect fatty acid biosynthesis. By constructing a network diagram of the DE-miRNAs and fatty acid biosynthesis, we found that hsa-miR-338-3p, hsa-miR-19b-3p, hsa-miR-15b-5p, hsa-miR-202-5p, hsa-miR-338-5p, hsa-miR-5011-5p, hsa-miR-3065-3p, hsa-miR-4528, hsa-miR-195-5p, hsa-miR-196a-5p, hsa-miR-483-3p, hsa-miR-19a-3p, hsa-miR-138-2-3p, hsa-miR-4530, hsa-miR-330-3p, and hsa-miR-362-5p may regulate fatty acid metabolism by regulating its predicted or validated target genes (Figure 3A). Among them, hsa-miR-4528, hsa-miR-19a-3p, hsa-miR-19b-3p, hsa-miR-195-5p, and hsa-miR-15b-5p were up-regulated in both blood and tissues of GBM, while hsa-miR-338-3p, hsa-miR-330-3p, hsa-miR-338-5p, and hsa-miR-138-2-3p were down-regulated both in blood and tissues of GBM, those miRNAs may regulate fatty acid metabolism by regulating its predicted or validated target genes (ACSL1, ACSL3, ACSL4, ACSL6, FASN, OXSM) (Figure 3B). GEPIA (http://gepia.cancer-pku.cn/detail.php) which can analyze RNAs from cancer genome atlas (TCGA) and genotype tissue expression project (GTEx) was used to analyze the differential expression of mRNA of ACSL1, ACSL3, ACSL4, ACSL6, FASN and OXSM in GBM and control tissues. OXSM is highly expressed in GBM (Figure 3C). High expression of OXSM in GBM suggests that OXSM may be involved in the mechanism of miRNAs regulating fatty acid metabolism.
Figure 3.
Fatty acid metabolism regulated by DE-miRNAs. (A) Network diagram of the DE-miRNAs and fatty acid biosynthesis produced by Cytoscape. (B) Network diagram of the upregulated miRNAs or downregulated miRNAs in all type of samples and their target genes in pathway of fatty acid biosynthesis produced by Cytoscape; (C) the expression levels of target genes in pathway of fatty acid biosynthesis from the GEPIA database. *P < 0.05.
Identification of OXSM as Direct Target of hsa-miR338-3p
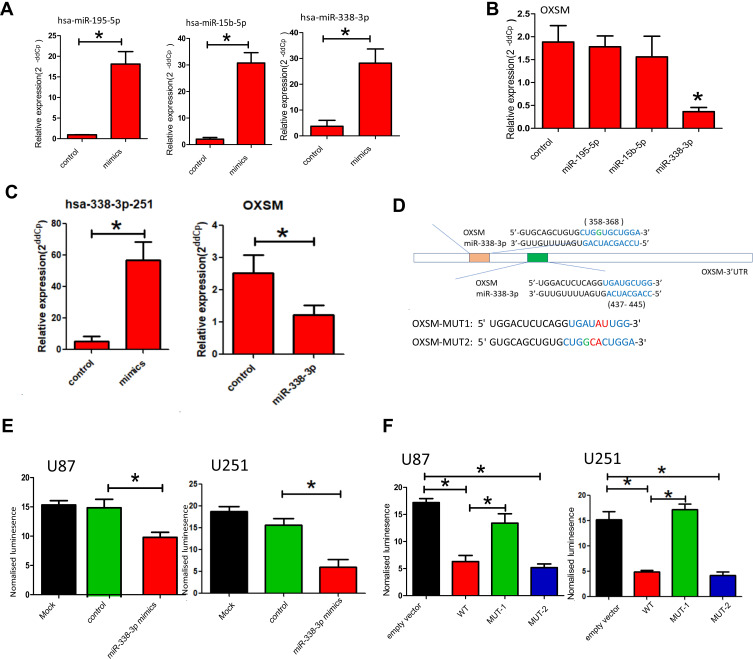
In our study, OXSM is a possible target gene predicted by 3 miRNAs (hsa-miR195-5p, hsa-miR15b-5p, and hsa-miR338-3p, Figure 4B). In order to verify the regulation of OXSM by these three miRNAs, we overexpressed the corresponding miRNA expression in the U87 cell lines by transfecting miRNA mimics (Figure 4A). Using qRT-PCR to detect the expression of OXSM, we found that hsa-miR338-3p can significantly inhibit the expression of OXSM both in U87 cell lines (Figure 4B). Similarly, hsa-miR338-3p down-regulated the expression level of OXSM in U251 cell lines.
Figure 4.
Identification of OXSM as direct target of hsa-miR338-3p. (A) hsa-miR195-5p, hsa-miR15b-5p and hsa-miR338-3p were overexpressed in the U87 cell lines by transfecting miRNA mimics; (B) the expression of OXSM in the U87 cell lines after transfecting of miRNA mimics; (C) hsa-miR338-3p were overexpressed in the U251 cell lines by transfecting miRNA mimics and the expression of OXSM in the U251 cell lines after transfecting of miRNA mimics; (D) the two putative binding sites between OXSM 3′UTR and hsa-miR338-3p; Luciferase report vectors carrying the full OXSM 3′UTR of mutation site1 (MUT-1, 437–445) and mutation site 2 (MUT-2, 358–368); (E) luminescence in hsa-miR338-3p-treated cells with wild-type reporters; (F) luminescence in hsa-miR338-3p-treated cells with MUT-1and MUT-2 reporters. *P < 0.05.
We explored the potential of hsa-miR338-3p to directly target the 3′UTR of OXSM. The two putative binding sites between OXSM 3′UTR and hsa-miR338-3p are shown in Figure 4D. Luciferase report vectors carrying the full 3′UTR of wild-type (WT) OXSM, mutation site1 (MUT-1, 437–445), or mutation site 2 (MUT-2, 358–368) were constructed (Figure 4D). Reporters were transfected into U87 and U251 cells with hsa-miR338-3p mimics, luminescence in hsa-miR338-3p-treated cells was clearly less than in controls (Figure 4E). The results show that hsa-miR338-3p can directly target the 3′UTR of OXSM and affect subsequent transcription. Therefore, we also worked to identify the effective binding site between OXSM and hsa-miR338-3p. No substantial differences in the inhibition luciferase activity between the WT reporter and mut-2 were observed, while the luciferase activity after transfection of the mut-1 reporter was significantly higher than that of the WT reporter both in U87 and U251 cell lines (Figure 4F). We found that hsa-miR338-3p can directly target OXSM 3′UTR through bind site 1 (437–445).
Hsa-miR338-3p Contribute to the Development of Glioma Cells via Expression of OXSM
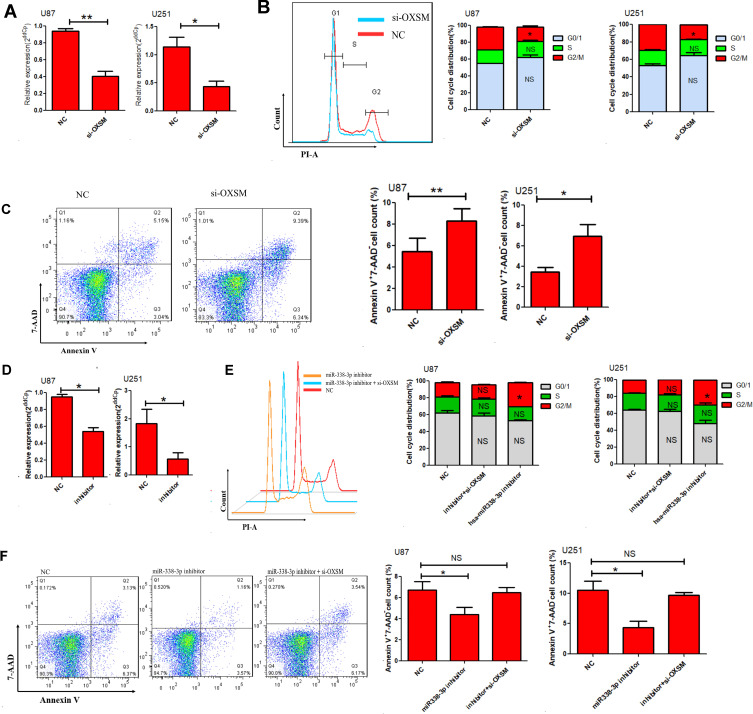
Omics studies have provided important information for understanding the pathogenesis of diseases. As an enzyme required for elongation of fatty acid chains in the mitochondria, we hypothesized that OXSM may contribute to the development of glioma cells by regulating the cell cycle and apoptosis. This was the first study addressing this question. Initially, we reduced the expression of OXSM in U87 and U251 cells by transfection with siRNA to investigate the effect of OXSM on cell cycle and apoptosis. After transfection (48 h), OXSM was low expressed (Figure 5A). We found that forced expression of OXSM significantly arrested U87 cell cycle (Figure 5B). Furthermore, the percentage of Annexin V+ 7-AAD− apoptotic cells was higher following the forced expression of OXSM (Figure 5C). These data indicate that OXSM promotes cell cycle and inhibits apoptosis of U87 and U251 cell lines.
Figure 5.
Hsa-miR338-3p regulate the cell cycle and apoptosis of glioma cells via OXSM. (A) The relative expression of OXSM was determined 48 h after transfection of 20 µM OXSM siRNA or controls through qRT-PCR. The cell cycle (B) and apoptosis (C) of GBM cells was determined after transfection of OXSM siRNA. (D) The expression of hsa-miR338-3p was inhibited by introducing 20 µM inhibitor to U87 and U251 cells. After transfection of hsa-miR338-3p inhibitor and OXSM siRNAs, the cell cycle (E) and apoptosis (F) of glioma cells were compared. *P < 0.05, **P < 0.01.
We subsequently assessed the role of hsa-miR338-3p in regulating the development of glioma cells by transfection of hsa-miR338-3p inhibitor into U87 and U251 cells. Following the reduced expression of hsa-miR338-3p (Figure 5D), U87 and U251 cells showed a significant increase in cell cycle (Figure 5E), and apoptosis was significantly reduced in comparison with the controls (Figure 5F). Furthermore, siRNA was used to suppress the expression of OXSM in hsa-miR338-3p -low-expressing U87 or U251 cells to verify that hsa-miR338 affects GBM cell development through regulation of OXSM. Following the downregulation of OXSM, there was no statistical difference detected in cell cycle or apoptosis compared with controls. These data suggest that hsa-miR338-3p inhibitors antagonize the effect of downregulation of OXSM on the function of GBM cells.
Discussion
It is believed that dysregulation of miRNAs is closely associated with the development of multiple human diseases in brain.21–24 During the past few years, studies have intensively suggested that DE-miRNAs and their downstream target genes are closely associated with the development of GBM.25–27 In this present study, we conducted a differential expression analysis using multiple public microarray datasets to investigate DE-miRNAs in blood and brain tissues of GBM. Upregulated DE-miRNAs and downregulated DE-miRNAs were finally identified in blood and brain tissues. Most of the DE-miRNAs we screened are consistent with the analysis results of previous studies. For example, hsa-miR-338-3p found to be significantly downregulated in GBM, acted as a tumor-suppressing gene whose silencing can inhibit malignant biological behaviors of glioma cells, was an independent prognostic biomarker associated with poor prognosis in glioma patients;28 hsa-miR-139-5p is lower in blood and brain tissues of GBM, was identified as a tumor suppressor by negatively targeting Notch1;29 hsa-miR-490-3p expression was significantly downregulated in GBM, which can inhibit glioma cell proliferation and migration.30
GO and KEGG pathway analysis offered insight into the possible roles of DE-miRNAs in the pathogenesis of GBM. We found that the IFN-gamma pathway was involved in the most significantly enriched terms shared by all sample types, indicates that DE-miRNAs in GBM may affect the activation of immune system and is a key factor affecting disease progression. Of note, in our study, “TRAIL signaling pathway” is one of 10 most significant pathways enriched for DE-miRNAs in blood and tissue. Type I interferon can activate DC cells to release tumor necrosis factor-related apoptosis-inducing ligand (TRAIL), thereby enhancing cytotoxicity of NK cells or directly killing tumor cells.31,32 “VEGF and VEGFR signaling network” were also significantly enriched for DE-miRNAs in our study. Vascular endothelial growth factor (VEGF) is one of the core members of tumor angiogenic factors. GBM has an increasingly strong angiogenic effect, and it is more dependent on neovascularization.33
In the past several years, a wealth of evidence has emerged on how metabolism affects aspects of biology. Considerable progress has been made in the field of cell metabolism, becoming one of the hottest areas in research.34,35 Metabolomics methods can establish a direct correlation between changes in metabolite content and changes in biological phenotypes.36,37 Similar to other types of cancers, fatty acid uptake and lipid metabolism are deregulated in malignant glioma.38,39 Among 10 most significantly KEGG pathways in our study, “Fatty acid biosynthesis” was identified for DE-miRNAs both in blood and tissue. The network relationship between DE-miRNA and fatty acid biosynthesis was further analyzed in this study and our results revealed that hsa-miR-338-3p may regulate fatty acid metabolism by directly targeting OXSM, an enzyme required for elongation of fatty acid chains in the mitochondria.
Emerging evidences suggest that cancer cells frequently reprogram metabolic pathways to meet their high demands of biogenesis and rapid proliferation. Dysregulation of metabolism ultimately influences cancer cell fate decision. Thus, understanding how key metabolic pathways, such as lipid metabolism, are aberrantly regulated, and what advantages these metabolic changes confer to cancer cells are of great interest and may benefit the follow-up research and therapeutic targeting. As an enzyme required for elongation of fatty acid chains, we assessed whether OXSM contributed to fate of GBM by regulating the function of glioma cells. In this study, the data revealed an accelerated cell cycle and an anti-apoptotic role for OXSM in U87 and U251 cells, which has not been reported. Our experiments show that OXSM, an enzyme required for elongation of fatty acid chains in the mitochondria, which is abnormally expressed in GBM, can not only participate in the regulation of fatty acid biosynthesis but also affect the biogenesis and rapid proliferation of glioma cells. hsa-miR-338-3p is abnormally expressed in various malignant tumors and participates in the proliferation, differentiation and invasion of tumor cells.40 It is evident that has-miR-338-3p is down-regulated in metastatic tumor tissues of neuroblastoma compared to primary tumors, and that has-miR-338-3p can inhibit cell proliferation by inducing cell cycle arrest, as well as restrain cell migration and invasion.41 Similarly, our data revealed the arrest-cell cycle and pro-apoptosis role of hsa-miR-338-3p in glioma cells. Considering that hsa-miR-338-3p can down-regulate the expression of the target gene OXSM, we also investigated whether hsa-miR-338-3p is involved in the regulation of U87 cell function by OXSM. This study confirmed that hsa-miR-338-3p inhibitor antagonized the effect of downregulation of OXSM on the function of U87 cells. Our results indicate that hsa-miR-338-3p may affect the biogenesis and rapid proliferation of glioma cells by regulating the level of OXSM. Current data have highlighted the importance of hsa-miR-338-3p and OXSM in GBM, which provide potential targets for improved immune intervention.
In summary, our analysis of microarray studies will facilitate the understanding of DE-miRNAs in GBM. The investigation identified hsa-miR-338-3p, down-regulated in GBM, plays an important metabolic regulatory role in GBM and may also affect the biogenesis and rapid proliferation of tumor cells by regulation the level of OXSM. This study provides useful information for the exploration of new intervention paths in GBM.
Acknowledgments
The authors thank all members of our lab for excellent technical help.
Abbreviations
OXSM, mitochondrial 3-oxoacyl-ACP synthase; GBM, glioblastoma; DE-miRNAs, miRNAs differentially expressed; OS, overall survival; miRNAs, microRNAs; TCGA, cancer genome atlas; GTEx, genotype tissue expression project; WT, wild-type; TRAIL, tumor necrosis factor-related apoptosis-inducing ligand; VEGF, vascular endothelial growth factor; EGFR, a receptor for members of the epidermal growth factor.
Data Sharing Statement
The data that support the findings of this study are available from University of California Santa Cruz Genome Browser and GEO database.
Ethics Approval
This work was approved by the Ethical Board of China Medical University.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Delgado-López PD, Corrales-García EM. Survival in glioblastoma: a review on the impact of treatment modalities. Clin Transl Oncol. 2016;18(11):1062–1071. doi: 10.1007/s12094-016-1497-x [DOI] [PubMed] [Google Scholar]
- 2.Weller M, van den Bent M, Hopkins K. EANO guideline for the diagnosis and treatment of anaplastic gliomas and glioblastoma. Lancet Oncol. 2014;15(9):e395–403. doi: 10.1016/S1470-2045(14)70011-7 [DOI] [PubMed] [Google Scholar]
- 3.Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised Phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466. doi: 10.1016/S1470-2045(09)70025-7 [DOI] [PubMed] [Google Scholar]
- 4.Yang Z, Zhang T, Wang Q, Gao H. Overexpression of microRNA-34a attenuates proliferation and induces apoptosis in pituitary adenoma cells via SOX7. Mol Ther Oncolytics. 2018;10:40–47. doi: 10.1016/j.omto.2018.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan R, Yang T, Zhai H, Zhou Z, Gao L, Li Y. MicroRNA-150-5p affects cell proliferation, apoptosis, and EMT by regulation of the BRAF V600E mutation in papillary thyroid cancer cells. J Cell Biochem. 2018;119(11):8763–8772. doi: 10.1002/jcb.27108 [DOI] [PubMed] [Google Scholar]
- 6.Xiao R, Li C, Chai B. miRNA-144 suppresses proliferation and migration of colorectal cancer cells through GSPT1. Biomed Pharmacother. 2015;74:138–144. doi: 10.1016/j.biopha.2015.08.006 [DOI] [PubMed] [Google Scholar]
- 7.Holmgren G, Synnergren J, Andersson CX, Lindahl A, Sartipy P. MicroRNAs as potential biomarkers for doxorubicin-induced cardiotoxicity. Toxicol in Vitro. 2016;34:26–34. doi: 10.1016/j.tiv.2016.03.009 [DOI] [PubMed] [Google Scholar]
- 8.Lin J, Chuang -C-C, Zuo L. Potential roles of microRNAs and ROS in colorectal cancer: diagnostic biomarkers and therapeutic targets. Oncotarget. 2017;8(10):17328–17346. doi: 10.18632/oncotarget.14461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qiu S, Lin S, Hu D, Feng Y, Tan Y, Peng Y. Interactions of miR-323/miR-326/miR-329 and miR-130a/miR-155/miR-210 as prognostic indicators for clinical outcome of glioblastoma patients. J Transl Med. 2013;11:10. doi: 10.1186/1479-5876-11-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen L, Sun C, Li Y. MicroRNA-199a-3p suppresses glioma cell proliferation by regulating the AKT/mTOR signaling pathway. Tumour Biol. 2015;36(9):6929–6938. doi: 10.1007/s13277-015-3409-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan Z, Wang J, Wang C, Jiao Y, Qi W, Che S. miR-96 /HBP1/Wnt/β-catenin regulatory circuitry promotes glioma growth. FEBS Lett. 2014;588(17):3038–3046. doi: 10.1016/j.febslet.2014.06.017 [DOI] [PubMed] [Google Scholar]
- 12.Shi Z-M, Wang X-F, Qian X. MiRNA-181b suppresses IGF-1R and functions as a tumor suppressor gene in gliomas. RNA. 2013;19(4):552–560. doi: 10.1261/rna.035972.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Z, Liu Y, Li L. MiR-7-5p is frequently downregulated in glioblastoma microvasculature and inhibits vascular endothelial cell proliferation by targeting RAF1. Tumour Biol. 2014;35(10):10177–10184. doi: 10.1007/s13277-014-2318-x [DOI] [PubMed] [Google Scholar]
- 14.Tanya Barrett 1, Dennis B, Stephen EW. NCBI GEO: archive for functional genomics data sets – 10 years on. Nucleic Acids Res. 2011;39:D1005–10. doi: 10.1093/nar/gkq1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marot G, Foulley J-L, Mayer C-D, Jaffrezic F. Moderated effect size and P-value combinations for microarray meta-analyses. Bioinformatics. 2009;25(20):2692–2699. doi: 10.1093/bioinformatics/btp444 [DOI] [PubMed] [Google Scholar]
- 16.Liang RF, Li M, Yang Y, Wang X, Mao Q, Liu YH. Circulating miR-128 as a potential diagnostic biomarker for glioma. Clin Neurol Neurosurg. 2017;160:88–91. doi: 10.1016/j.clineuro.2017.06.020 [DOI] [PubMed] [Google Scholar]
- 17.Sun J, Shi H, Lai N, Liao K, Zhang S, Lu X. Overexpression of microRNA-155 predicts poor prognosis in glioma patients. Med Oncol. 2014;31(4):911. doi: 10.1007/s12032-014-0911-x [DOI] [PubMed] [Google Scholar]
- 18.Yan Z, Che S, Wang J, Jiao Y, Wang C, Meng Q. miR-155 contributes to the progression of glioma by enhancing Wnt/β-catenin pathway. Tumour Biol. 2015;36(7):5323–5331. doi: 10.1007/s13277-015-3193-9 [DOI] [PubMed] [Google Scholar]
- 19.Liu Q, Zou R, Zhou R. miR-155 regulates glioma cells invasion and chemosensitivity by p38 isforms in vitro. J Cell Biochem. 2015;116(7):1213–1221. doi: 10.1002/jcb.25073 [DOI] [PubMed] [Google Scholar]
- 20.Kannan R, Lyon I, Baker N. Dietary control of lipogenesis in vivo in host tissues and tumors of mice bearing Ehrlich ascites carcinoma. Cancer Res. 1980;40(12):4606–4611. [PubMed] [Google Scholar]
- 21.Hara N, Kikuchi M, Miyashita A. Serum microRNA miR-501-3p as a potential biomarker related to the progression of Alzheimer’s disease. Acta Neuropathol Commun. 2017;5(1):10. doi: 10.1186/s40478-017-0414-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bekris LM, Lutz F, Montine TJ. MicroRNA in Alzheimer’s disease: an exploratory study in brain, cerebrospinal fluid and plasma. Biomarkers. 2013;18(5):455–466. doi: 10.3109/1354750X.2013.814073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng S-J, Zhang X-Q, Li J-T, Dai X-M, Zhao F. miRNA-223 regulates ischemic neuronal injury by targeting the type 1 insulin-like growth factor receptor (IGF1R). Folia Neuropathol. 2018;56(1):49–57. doi: 10.5114/fn.2018.74659 [DOI] [PubMed] [Google Scholar]
- 24.Adlakha YK, Saini N. Brain microRNAs and insights into biological functions and therapeutic potential of brain enriched miRNA-128. Mol Cancer. 2014;13:33. doi: 10.1186/1476-4598-13-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Møller HG, Rasmussen AP, Andersen HH, Johnsen KB, Henriksen M, Duroux M. A systematic review of microRNA in glioblastoma multiforme: micro-modulators in the mesenchymal mode of migration and invasion. Mol Neurobiol. 2013;47(1):131–144. doi: 10.1007/s12035-012-8349-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hui X, Zhang S, Wang Y. miR‑454‑3p suppresses cell migration and invasion by targeting CPEB1 in human glioblastoma. Mol Med Rep. 2018;18(4):3965–3972. doi: 10.3892/mmr.2018.9386 [DOI] [PubMed] [Google Scholar]
- 27.Mao Y, Wei F, Wei C, Wei C. microRNA‑574 inhibits cell proliferation and invasion in glioblastoma multiforme by directly targeting zinc finger E‑box‑binding homeobox 1. Mol Med Rep. 2018;18(2):1826–1834. doi: 10.3892/mmr.2018.9106 [DOI] [PubMed] [Google Scholar]
- 28.Shang C, Hong Y, Guo Y, Xue Y-X. Mir-338-3p inhibits malignant biological behaviors of glioma cells by targeting MACC1 gene. Med Sci Monit. 2016;22:710–716. doi: 10.12659/msm.897055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J, Li Q, Lin L. Targeting the Notch1 oncogene by miR-139-5p inhibits glioma metastasis and epithelial-mesenchymal transition (EMT). BMC Neurol. 2018;18(1):133. doi: 10.1186/s12883-018-1139-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang F, Wu A, Wang Y, Liu J. miR-490-3p functions as a tumor suppressor in glioma by inhibiting high-mobility group AT-hook 2 expression. Exp Ther Med. 2019;18(1):664–670. doi: 10.3892/etm.2019.7606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Demoulin S, Herfs M, Delvenne P, Hubert P. Tumor microenvironment converts plasmacytoid dendritic cells into immunosuppressive/tolerogenic cells: insight into the molecular mechanisms. J Leukoc Biol. 2013;93(3):343–352. doi: 10.1189/jlb.0812397 [DOI] [PubMed] [Google Scholar]
- 32.Dunn GP, Bruce AT, Sheehan KC, et al. A critical function for type I interferons in cancer immunoediting. Nat Immunol. 2005;6(7):722–729. doi: 10.1038/ni1213 [DOI] [PubMed] [Google Scholar]
- 33.Folkman J, Merler E, Abernathy C, Williams G. Isolation of a tumor factor responsible for angiogenesis. J Exp Med. 1971;133(2):275–288. doi: 10.1084/jem.133.2.275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Sullivan D, Pearce EL. Targeting T cell metabolism for therapy. Trends Immunol. 2015;36(2):71–80. doi: 10.1016/j.it.2014.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kahlert UD, Mooney SM, Natsumeda M, et al. Targeting cancer stem-like cells in glioblastoma and colorectal cancer through metabolic pathways. Int J Cancer. 2017;140(1):10–22. doi: 10.1002/ijc.30259 [DOI] [PubMed] [Google Scholar]
- 36.Nobeli I, Thornton JM. A bioinformatician’s view of the metabolome. BioEssays. 2006;28(5):534–545. doi: 10.1002/bies.20414 [DOI] [PubMed] [Google Scholar]
- 37.Benjamin D, Cravatt B, Nomura D. Global profiling strategies for mapping dysregulated metabolic pathways in cancer. Cell Metab. 2012;16(5):565–577. doi: 10.1016/j.cmet.2012.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng C, Geng F, Cheng X, et al. Lipid metabolism reprogramming and its potential targets in cancer. Cancer Commun. 2018;38(1):27. doi: 10.1186/s40880-018-0301-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ray U, Roy SS. Aberrant lipid metabolism in cancer cells – the role of oncolipid-activated signaling. FEBS J. 2018;285(3):432–443. doi: 10.1111/febs.14281 [DOI] [PubMed] [Google Scholar]
- 40.Sui GQ, Fei D, Guo F, et al. MicroRNA-338-3p inhibits thyroid cancer progression through targeting AKT3. Am J Cancer Res. 2017;7(5):1177–1187. [PMC free article] [PubMed] [Google Scholar]
- 41.Chen X, Pan M, Han L, Lu H, Hao X, Dong Q. miR-338-3p suppresses neuroblastoma proliferation, invasion and migration through targeting PREX2a. FEBS Lett. 2013;587(22):3729–3737. doi: 10.1016/j.febslet.2013.09.044 [DOI] [PubMed] [Google Scholar]