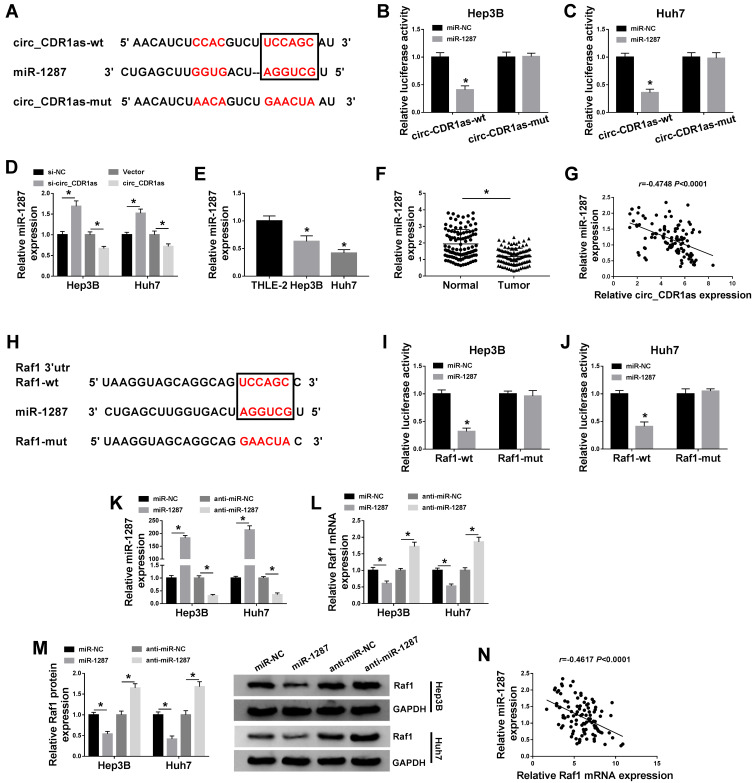
Figure 5.
Circ_CDR1as targets miR-1287, an upstream miRNA of Raf1. (A) The binding sites of miR-1287 on circ_CDR1as, as well as the mutant sites based on starBase. (B and C) Dual-luciferase reporter assay for the luciferase activities of circ_CDR1as-wt and circ_CDR1as-mut in Hep3B and Huh7 cells. (D) QRT-PCR assay for the relative expression of miR-1287 in Hep3B and Huh7 cells introduced with si-NC, si-circ_CDR1as, Vector or circ_CDR1as. (E) QRT-PCR assay the relative expression of miR-1287 in THLE-2, Hep3B and Huh7 cells. (F) QRT-PCR assay for the relative expression of miR-1287 in HCC tissues and adjacent normal tissues (n=105). (G) Pearson correlation analysis for the correlation between the levels of miR-1287 and circ_CDR1as in HCC tissues, r=−0.4748, P < 0.0001. (H) The binding sites of miR-1287 on the 3ʹUTR of Raf1, as well as the mutant sites based on starBase. (I and J) Dual-luciferase reporter assay for the luciferase activities of Raf1-wt and Raf1-mut in Hep3B and Huh7 cells. (K–M) Hep3B and Huh7 cells are transfected with miR-NC, miR-1287, anti-miR-NC or anti-miR-1287. (K and L) QRT-PCR assay the relative expression levels of miR-1287 and Raf1 in transfected cells. (M) Western blot analysis for the protein level of Raf1 in transfected cells. (N) Pearson correlation analysis for the correlation between the levels of miR-1287 and Raf1 in HCC tissues, r=−0.5270, P < 0.0001. *P < 0.05.