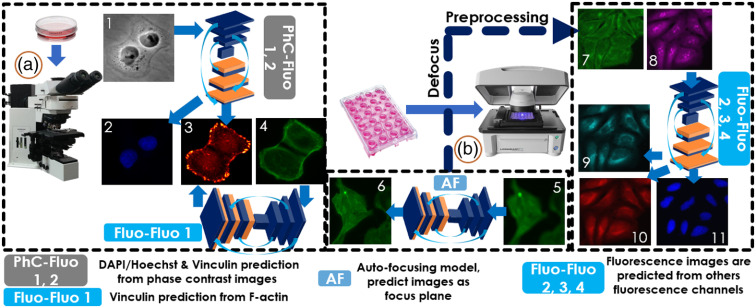
Fig. 1.
Virtual fluorescence imaging pipeline for cell microscopy with all models implemented in our study. MDA-MB-231 cells (on the left) were imaged to collect co-registered phase contrast, DAPI, F-actin, and vinculin. The PhC-Fluo models were trained to predict fluorescence images from phase contrast (either DAPI or vinculin). The Fluo-Fluo models using MDA-MB-231 in Fluo-Fluo model 1 and U2OS-CPS in Fluo-Fluo models 2, 3, 4 (on the right) predicted florescence images from a co-registered fluorescence channel, from the same cells. Although each model performs different goals in our study, out-of-focus images were fed to the AF model (at center) as a preprocessing step to refocus the cell images (U2OS-AF) before reuse in training and testing of the Fluo-Fluo models. (1) Phase contrast, (2) DAPI, (3) vinculin, (4) F-actin, (5) out-of-focus F-actin, (6) in-focus predicted Factin, (7) Golgi apparatus plasma membrane F-actin, (8) nucleoli and cytoplasmic RNA, (9) endoplasmic reticulum, (10) mitochondria, and (11) DAPI/Hoechst. (a) Regular fluorescence imaging for MDA-MB-231 cells collection and (b) automated cellular imaging system for U2OS-CPS/AF data collection. Details of sample preparation are described in Sec. 2.2.