Figure 1.
CFAV-Derived Endogenous Viral Elements Interact with Natural CFAV Infection through the piRNA Pathway
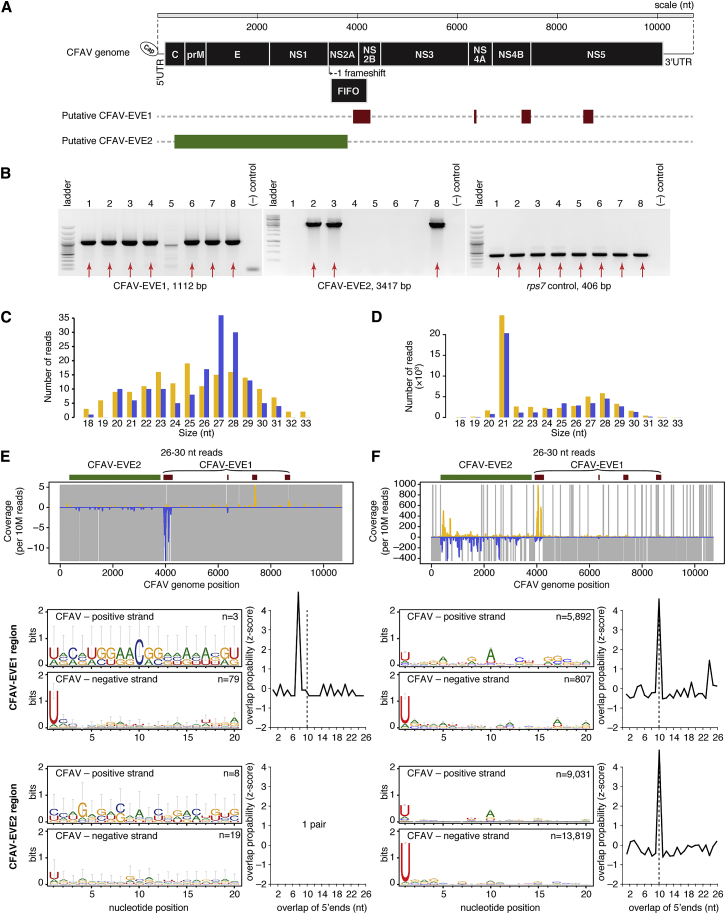
(A) The schematic represents two potential CFAV EVE structures detected in publicly available Ae. aegypti sequences.
(B) The presence of putative CFAV-EVE1 and CFAV-EVE2 in eight mosquitoes from the same outbred colony was verified by PCR with primers specific to CFAV-EVE1 (left), CFAV-EVE2 (middle), and rps7 gene control (right).
(C and D) Size distribution of sRNAs mapping to the CFAV genome from naturally CFAV-uninfected (C) and CFAV-infected (D) mosquitoes from the outbred colony.
(E and F) Analysis of CFAV-derived piRNAs from naturally CFAV-uninfected (E) and CFAV-infected (F) mosquitoes from the outbred colony. Mapping of 26- to 30-nt sRNAs (top), sequence logos of 26- to 30-nt sRNAs (bottom left), and overlap probability of 26- to 30-nt sRNAs (bottom right) is shown. Sequence logos and overlap probability for CFAV-EVE1 were restricted to the NS2 region.
In (C)–(F), positive- and negative-sense reads with respect to the reference CFAV genome are shown in yellow and blue, respectively. Uncovered nucleotides are represented by gray lines. See also Data S1, Figure S1, Table S1, Table S2, and Table S5.