Figure 2.
CFAV-EVE1 Interacts with Experimental CFAV Infection through the piRNA Pathway
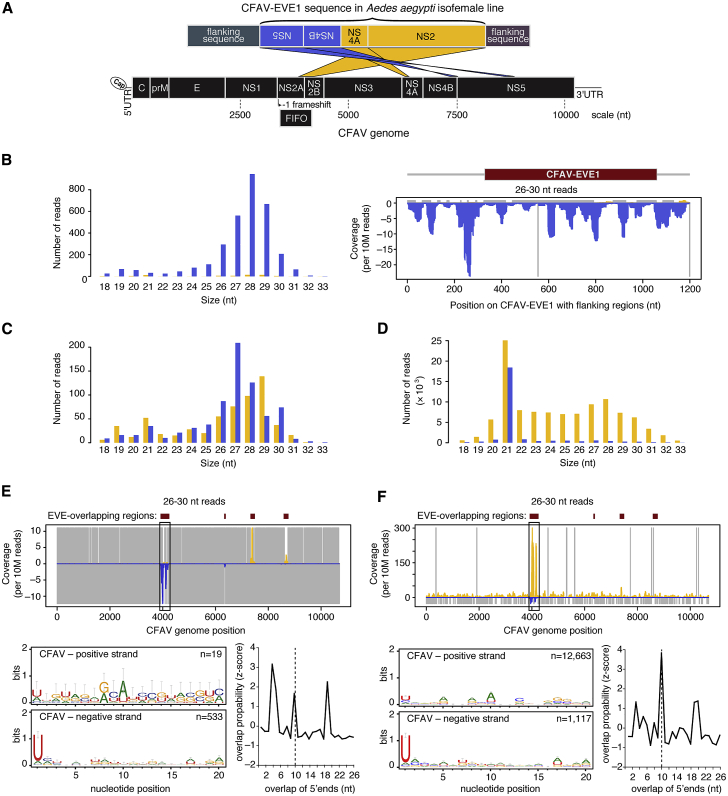
(A) Schematic of the CFAV-EVE1 structure in the CFAV-free isofemale line represented as the alignment of the EVE locus in the Ae. aegypti genome assembly AaegL3 (top) to the genome of the CFAV-KPP isolate (bottom). CFAV-EVE1 comprises four different regions of the CFAV genome. Yellow and blue colors indicate forward and reverse strands, respectively, according to the transcription direction in the supercontig.
(B) Production of piRNAs from CFAV-EVE1 in the CFAV-free isofemale line, represented as the size distribution (left) and alignment to the CFAV-EVE-1 locus (right). Blue color corresponds to negative-sense reads with respect to the mapping reference.
(C and D) Size distribution of sRNAs mapping to the CFAV genome from experimentally CFAV-uninfected (C) and CFAV-infected (D) mosquitoes from the isofemale line.
(E and F) Analysis of CFAV-derived piRNAs from experimentally CFAV-uninfected (E) and CFAV-infected (F) mosquitoes from the isofemale line. Mapping of 26- to 30-nt sRNAs (top), sequence logos of 26- to 30-nt sRNAs (bottom left), and overlap probability of 26- to 30-nt sRNAs (bottom right) is shown. Sequence logos and overlap probability were restricted to the NS2 region.
In (C)–(F), positive- and negative-sense reads with respect to the reference CFAV genome are shown in yellow and blue, respectively. Uncovered nucleotides are represented by gray lines. See also Figure S2 and Table S5.