Figure 4.
Ablation of CFAV-EVE1 Prevents CFAV-Derived piRNA Amplification
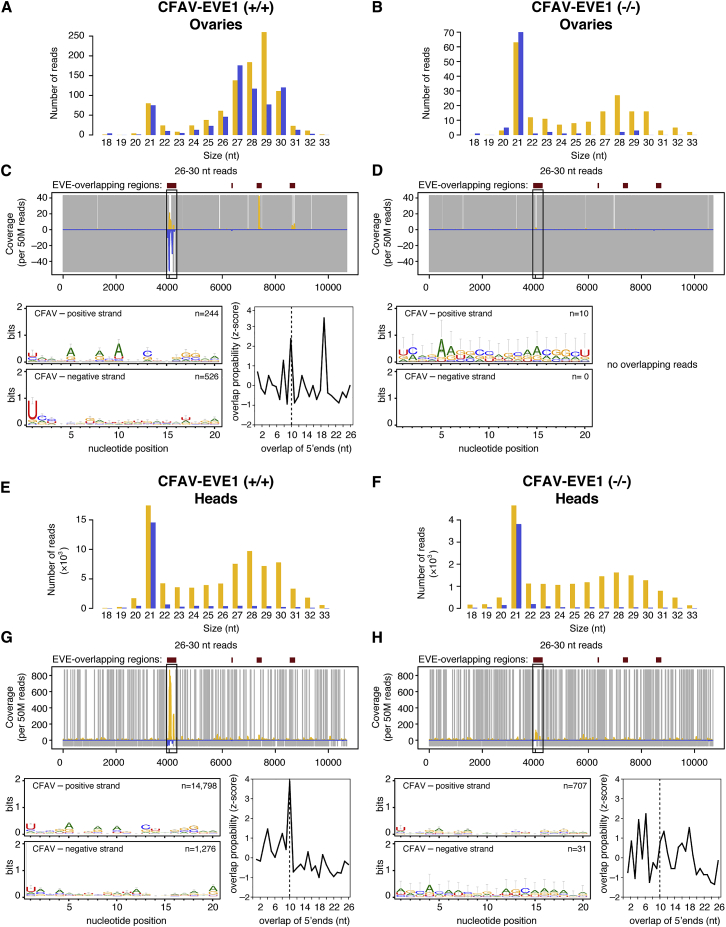
(A, B, E, and F) Size distribution of sRNAs mapping to the CFAV genome in ovaries (A and B) and heads (E and F) from experimentally infected CFAV-EVE1 (+/+) (A and E) and CFAV-EVE1 (−/−) (B and F) mosquitoes 7 days post-injection.
(C, D, G, and H) Analysis of CFAV-derived piRNAs in ovaries (C and D) and heads (G and H) from experimentally infected CFAV-EVE1 (+/+) (C and G) and CFAV-EVE1 (−/−) (D and H) mosquitoes 7 days post-injection. Mapping of 26- to 30-nt sRNAs (top), sequence logos of 26- to 30-nt sRNAs (bottom left), and overlap probability of 26- to 30-nt sRNAs (bottom right) is shown. Sequence logos and overlap probability were restricted to the NS2 region. In all panels, positive- and negative-sense reads with respect to the reference CFAV genome are shown in yellow and blue, respectively. Uncovered nucleotides are represented by gray lines.
See also Figures S3, S4 and Table S5.