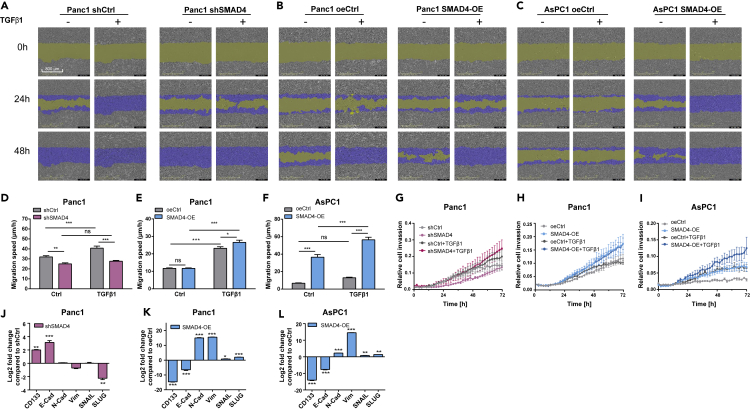
Figure 5.
Migration and Invasion Analysis of SMAD4-KD and SMAD4-OE PDA Cells With or Without TGFβ1 Stimulations and Corresponding Expression Levels of EMT Markers
(A–C) Migration assays were performed with the IncuCyte S3 Live Cell System Analysis using a scratch wound assay. Partial representation of the scratch wound assay for SMAD4-KD (A) or SMAD4-OE (B, C) and their control (shCtrl and oeCtrl) conditions in PDA cells with additional TGFβ1 or its solvent stimulation at 0, 24, 48 h. Images were obtained with the IncuCyte S3 Software. Yellow mask indicates the wound boundaries. Blue mask indicates the initial scratch wound area.
(D–F) Average cell migration speed distribution in Panc1 (D, E) and AsPC1 (F) cells containing SMAD4-KD or SMAD4-OE constructs and their empty vectors (shCtrl and oeCtrl) with additional TGFβ1 or its solvent stimulation, (n ≥ 8, mean ± SEM; two-way ANOVA, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001)..
(G–I) Invasion assays were carried out using a chemotaxis cell invasion assay for the IncuCyte S3 Live Cell System Analysis. Trans-well invasion assays for SMAD4-KD (G) or SMAD4-OE (H, I) and their control (shCtrl and oeCtrl) conditions in PDA cells were measured every 2h. Quantification was performed by measuring the total phase area of the bottom layer of the inner chamber within 72 h. Data were normalized for the initial value (n ≥ 6, mean ± SEM).
(J–L) Expression levels of EMT and cancer stemness-related genes (E-cadherin, N-cadherin, Vimentin, SNAIL, SLUG,and CD133) in SMAD4-KD (J) or SMAD4-OE ( K, L) conditions compared with their control conditions, shCtrl and oe-Ctrl respectively (n = 3, mean ± SEM, t test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
See also Figure S7.