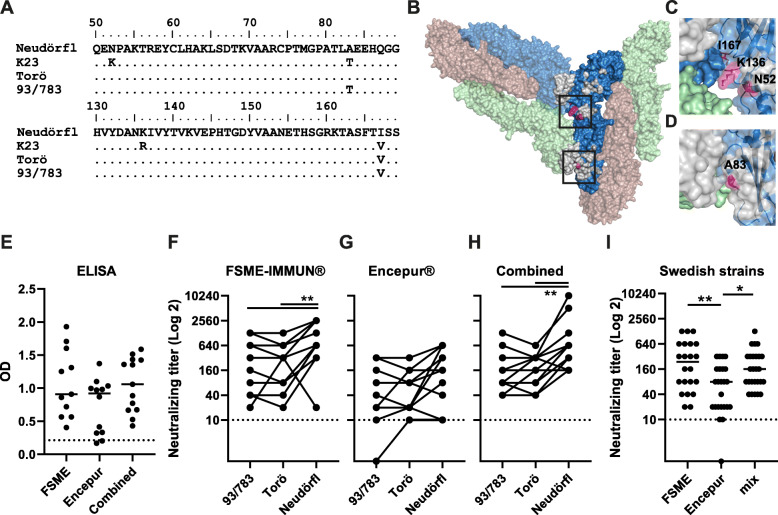
Fig. 6.
Sequence alignment. Structure of the E protein tries to explain the differences in neutralizing ability between the two vaccines. a Sequence alignment of the two vaccine TBEV strains Neudörfl (FSME-IMMUN®, reference) and K23 (Encepur®) with 93/783 and Torö using BioEdit software. b Two asymmetric units of the TBEV virion. Proposed antibody interacting sites [42] marked in grey and amino acids variations within the sites highlighted in pink. Magnification of neutralizing antibody interacting site close to the 3-fold symmetry axis (c) with the amino acid N52, K136, and I167 shown in pink, and the 5-fold symmetry axis (d) with the amino acid A83 shown in pink. PDB accession number 5O6A. Molecular graphics and structure editing were prepared using UCSF Chimera (https://www.cgl.ucsf.edu/chimera/) and PyMol (https://pymol.org/2/). e Anti-TBEV IgG ELISA of sera from individuals vaccinated with either FSME-IMMUN® (no. 11), Encepur® (no. 12) or a combination of both vaccines (no. 13), threshold set to < 0.213 dotted line. Neutralizing antibody titers against 93/783, Torö, and Neudörfl from individuals vaccinated with either FSME-IMMUN® (f), Encepur® (g), or a combination of both vaccines (h). Dotted line indicates threshold for neutralizing antibody titers. i Comparison of neutralizing antibody titers against 93/783 and Torö in vaccinated individuals with the different vaccines. Statistical significance was calculated using Friedman test (f, h) and Kruskal-Wallis multiple comparison test (i)