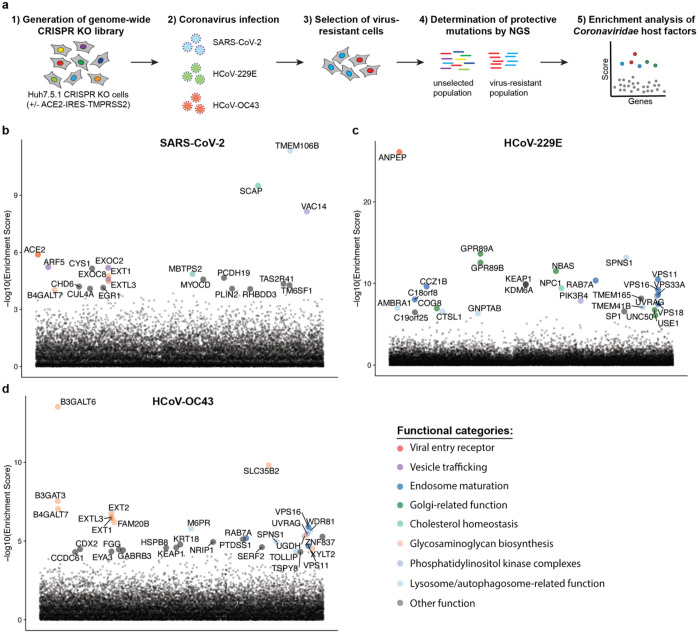
Figure 1: Genome-wide CRISPR KO screens in human cells identify host factors important for infection by for SARS-CoV-2, HCoV-229E and HCoV-OC43.
(a) Schematic of CRISPR KO screens for the identification of coronavirus host factors. Huh7.5.1-Cas9 (with bicistronic ACE2-IRES-TMPRSS2 construct for SARS-CoV-2 and without for 229E and OC43 screen) were mutagenized using a genome-wide sgRNA library. Mutant cells were infected with each coronavirus separately and virus-resistant cells were harvested 10–14 days post infection (dpi). The abundance of each sgRNA in the starting and selected population was determined by high-throughput sequencing and a gene enrichment analysis was performed. (b–d) Gene enrichment of CRISPR screens for (b) SARS-CoV-2, (c) 229E and (d) OC43 infection. Enrichment scores were determined by MaGECK analysis and genes were colored by biological function. The SARS-CoV-2 was performed once. The 229E and OC43 screens were performed twice and combined MaGECK scores are displayed.