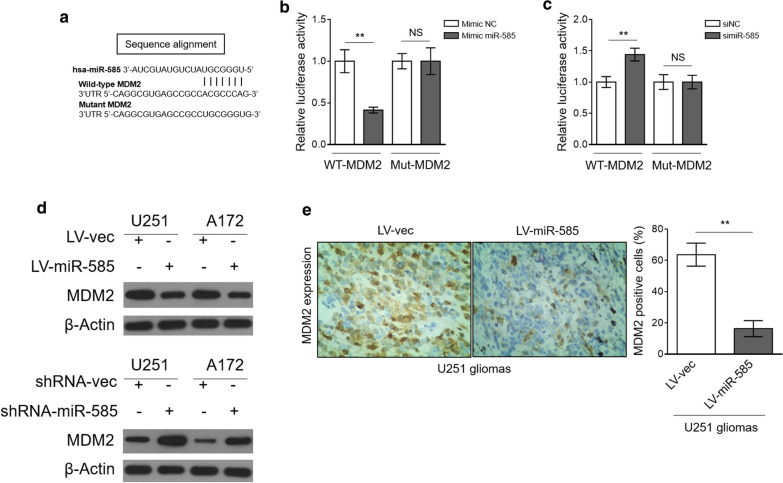
Fig. 4.
miR-585 directly targets MDM2. a The putative binding site of miR-585 in 3′-UTR of MDM2. b HEK293 cells were cotransfected mimic negative control (NC) or mimic miR-585 with firefly luciferase reporter containing wild type (WT) or mutant (Mut) 3′-UTR of MDM2. The luciferase activity was determined by dual-luciferase reporter assay. Data are mean ± SD (n = 3). Two-tailed unpaired Student’s t-test. **P < 0.01. c HEK293 cells were cotransfected siRNA negative control (siNC) or siRNA miR-585 with firefly luciferase reporter containing WT or Mut 3′-UTR of MDM2. The luciferase activity was determined by dual-luciferase reporter assay. Data are mean ± SD (n = 3). Two-tailed unpaired Student’s t-test. **P < 0.01. d The protein expression of MDM2 in U251 and A172 cells with miR-585 overexpression (upper) or knockdown (lower) was determined by Western blot analysis. β-Actin was used as a loading control. The representative images from 3 independent experiments are shown. e MDM2 expression in U251 tumor sections from Fig. 3d was analyzed by IHC staining. The representative images (left) and quantification analysis of MDM2 expression (right) are shown. Twelve random fields from 3 sections of each group were quantified. Data are mean ± SD (n = 3). Two-tailed unpaired Student’s t-test. **P < 0.01