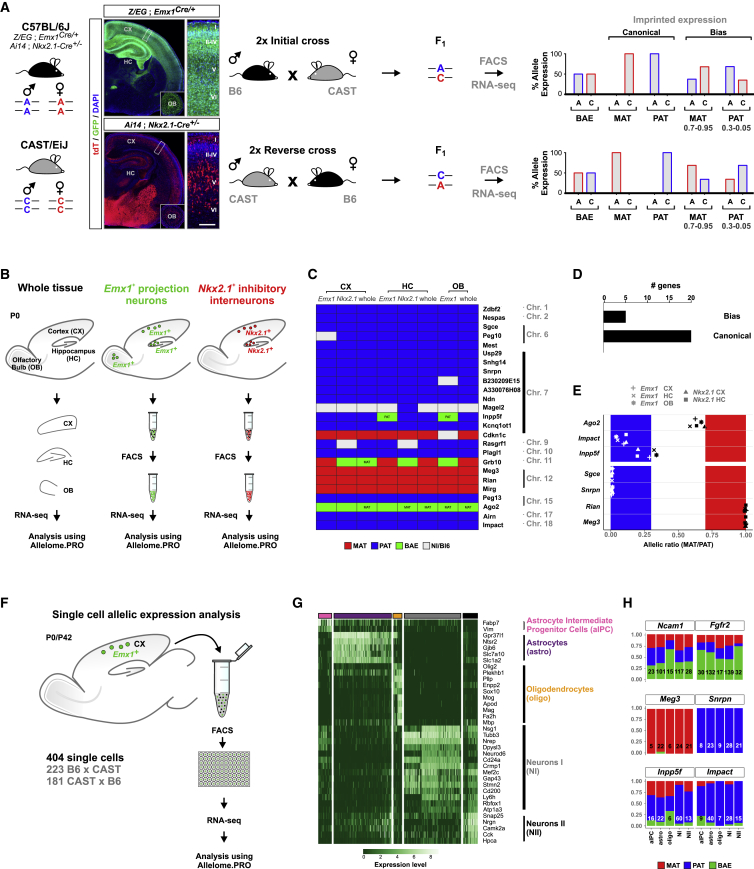
Figure 1.
Uniform Allelic Expression of Imprinted Genes in Major Forebrain Cell Types
(A) Strategy for cell-type-specific allelic expression analysis. Left: overview of parental Z/EG;Emx1Cre/+ and Ai14;Nkx2.1-Cre+/− reporter in a B6 genetic background and CAST mouse strains. Images depict neocortex (CX), hippocampus (HC), and OB (insets) in experimental B6/CAST mice with labeling of Emx1+ (Z/EG;Emx1Cre/+) and Nkx2.1+ (Ai14;Nkx2.1-Cre+/−) cell lineages in overview and at higher magnification (boxed areas in overview) at P0. GFP expression (green) and tdT expression (red) are indicated. Nuclei were stained using DAPI (blue). Cortical layers are indicated (roman numerals). Scale bar: overview, 500 μm; magnification, 60 μm; OB insets, 600 μm. Middle: breeding scheme for generating F1 B6xCAST hybrids with expected SNPs in F1. Right: expected relative allelic SNP expression upon FACS and RNA-seq analysis for biallelically expressed genes (BAE), canonical imprinted genes, and genes with expression bias.
(B) Experimental strategy for the analysis of allelic expression in bulk samples from whole tissue, Emx1+, and Nkx2.1+ lineages from CX, HC, and OB at P0 using the Allelome.PRO pipeline.
(C) Heatmap showing allelic expression of 25 known imprinted genes in whole tissue (whole), Emx1+, and Nkx2.1+ cell types in CX, HC, and OB. mat (red); pat (blue); BAE (green); NI/B6, not informative or expression bias toward B6 allele (white). The mat and pat labeling within individual boxes indicates genes with consistently higher expression in that direction below the allelic ratio cutoff (0.7).
(D) Number of genes in (C) with biased expression or canonical imprinting.
(E) Allelic ratio (mat/pat) of genes with canonical imprinting (Sgce, Snrpn, Rian, and Meg3) or biased expression (Impact, Ago2, and Inpp5f).
(F) Experimental strategy for allelic expression analysis in single cells of Emx1+ lineage at P0 and P42 using the Allelome.PRO pipeline. Numbers of cells analyzed from B6xCAST and reciprocal CASTxB6 crosses are indicated.
(G) Heatmap displays expression of a representative set of marker genes for classification of individual cell types in the Emx1+ lineage. Cell types (columns) and genes (rows) were ordered arbitrarily after hierarchical clustering. Colored bars above the heatmap indicate different cell types: aIPC (pink), astros (purple), oligos (orange), NI (gray), NII (black).
(H) Allelic expression of selected genes in single cells in defined cell types: Ncam1 and Fgfr2, biallelic expression; Meg3, canonical mat; Snrpn, canonical pat; Inpp5f and Impact, biased pat. Numbers indicate informative cells.
See also Figure S1.