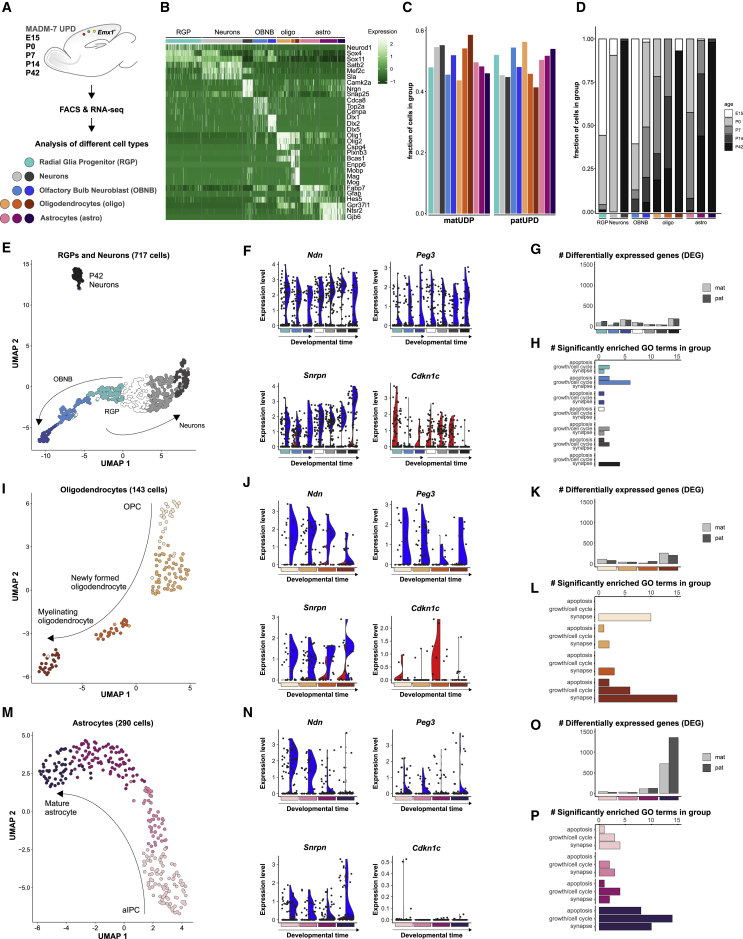
Figure 4.
Developmental Time Course Analysis of chr7 UPD Single-Cell Transcriptomes
(A) Experimental outline for analysis of single-cell transcriptomes in the Emx1+ lineage with MADM-induced UPD of chr7 at E15, P0, P7, P14, and P42.
(B) Heatmap displays expression of a representative set of marker genes for the classification of individual cell types in the Emx1+ lineage. Colored bars above the heatmap indicate different cell types: RGPs (cyan), neurons (light and dark gray), OBNBs (light and dark blue), oligos (orange), astros (purple).
(C) Fractions of cells with matUPD and patUPD in distinct cell types.
(D) Age distribution (E15, white; P0, light gray; P7, gray; P14, dark gray; P42, black) of analyzed cells indicated as the relative fraction in distinct cell types.
(E–P) Re-clustering of RGPs, neuronal cells, oligos, and astros. (E, I, and M) UMAP dots indicate individual cells. Neuronal cells (n = 717) were separated into 7 developmental clusters; RGP, OBNB, and neuronal classes (E). oligos (n = 143) were separated into 4 developmental clusters; oligodendrocyte progenitor cells (OPCs) and newly formed and myelinating oligos (I). astros (n = 290) were separated into 4 developmental clusters; aIPCs, immature astros, and mature astros (M). See also Figure S5. (F, J, and N) Violin plots show distribution of imprinted gene expression (Ndn, Peg3, Snrpn: paternally expressed; Cdkn1c: maternally expressed) in single cells (black dots) with matUPD (red, left side) and patUPD (blue, right side) from neurons (F), oligodendrocytes (J), and astrocytes (N). (G, K, and O) Number of DEGs in matUPD and patUPD at defined developmental stages (padj < 0.2, likelihood-ratio test) in neurons (G), oligodendrocytes (K), and astrocytes (O). (H, L, and P) Number of significantly enriched GO terms (p < 0.01, hypergeometric test) in apoptosis, growth/cell cycle, and synapse groups in neurons (H), oligodendrocytes (L), and astrocytes (P).