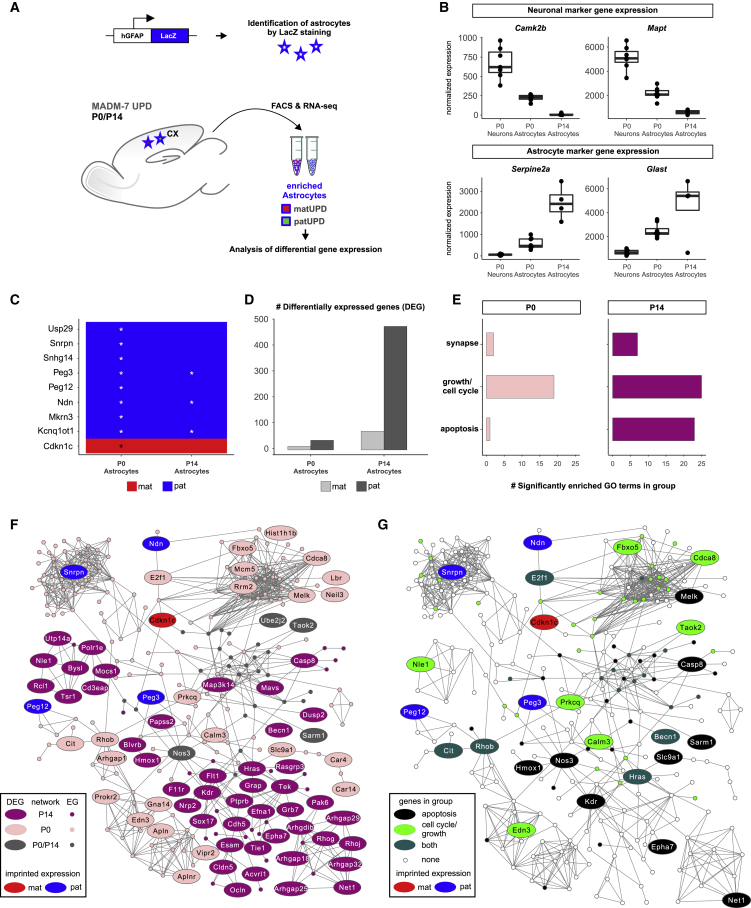
Figure 6.
Transcriptome Analysis in Cortical Astrocytes with chr7 UPD Reveals Deregulated Gene Networks Modulating Growth and Apoptosis
(A) Strategy to FACS-enrich cortical astrocytes with chr7 UPD using hGFAP promoter-driven LacZ transgene at P0 and P14 for RNA-seq and differential gene expression analysis.
(B) Normalized expression of marker genes for neurons (Camk2b and Mapt) and astrocytes (Serpine2a and Glast).
(C) Heatmap showing differential expression of 9 imprinted genes in matUPD/patUPD astrocytes at P0 and P14. Genes with higher expression in matUPD cells (log2 fold change > 0) are marked in red, and genes with higher expression in patUPD cells (log2 fold change < 0) are marked in blue. Asterisks mark significant differential expression (padj < 0.05, DESeq2).
(D) Number of DEGs (padj < 0.1, DESeq2) in astrocytes with matUPD (light gray) or patUPD (dark gray) at P0 and P14.
(E) Number of significantly enriched GO terms (p < 0.1, hypergeometric test) related to apoptosis, growth/cell cycle, and synapse.
(F and G) Merged top significant gene subnetworks identified by PhenomeExpress (p = 0.001) at P0/P14 that associate with deregulated imprinted genes in astrocytes with chr7 UPD. Each circle represents an individual gene. Imprinted genes are in red (mat) and blue (pat).
(F) Genes detected in P0, P14, or both subnetworks are in pink, purple, or gray, respectively. Large circles indicate significant deregulation (padj < 0.1, DESeq2, DEG) at P0 and/or P14. Small circles indicate expressed genes (EGs).
(G) Deregulated gene subnetworks highlighting genes involved in apoptosis (black), cell cycle/growth (green), or both (dark green).