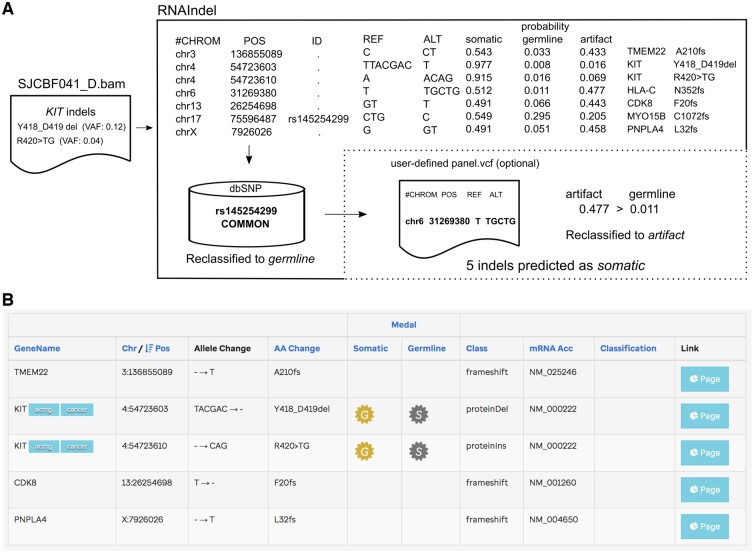
Fig. 3.
Pathogenic indel discovery by RNAIndel. (A) An example workflow is shown with a leukemia sample SJCBF041_D which harbors two distinct subclonal indels in the KIT oncogene based on WGS and WES analysis: Y418_D419del (VAF: 0.12) and R420>TG (0.04). Of the 7 somatic indels predicted by RNAIndel, only five remains after the refinement step. (B) By uploading the output VCF file to St Jude Cloud tool PeCanPIE, users can prioritize indels by pathogenicity. The KIT indels in somatic context were prioritized with the highest pathogenicity rank ‘gold’ (the ‘G’ symbol)