Fig. 4.
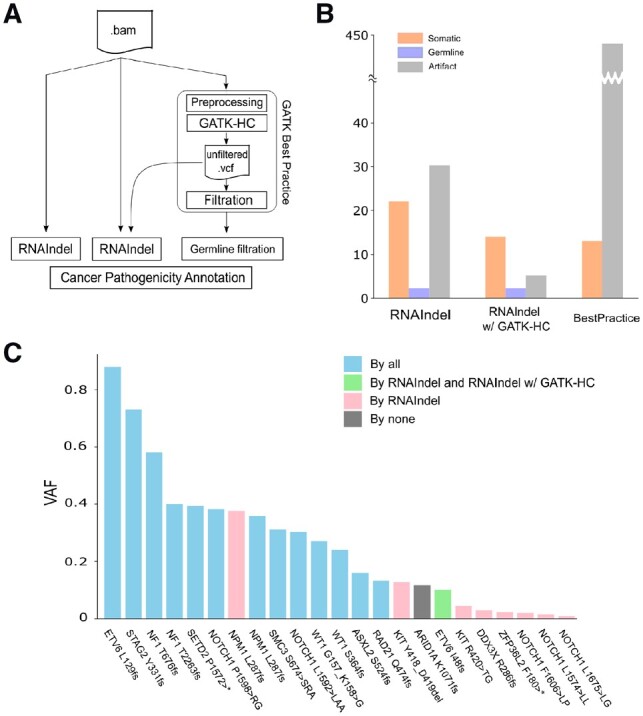
Working with an external variant caller. RNAIndel is flexibly designed to work with an external variant caller via a user-provided VCF file. GATK-HC was used as an example. (A) TestSet 1 (n = 77) was screened for pathogenic somatic indels by three approaches: 1) RNAIndel with built-in caller (left) using a RNA-Seq BAM file as input; 2) RNAIndel with GATK-HC (middle) using the VCF file from GATK-HC and the RNA-Seq BAM file as input; and 3) Best Practice-based approach (right). (B) Comparison of predicted somatic indels by the three approaches with the truth datasets. (C) Performance of the three methods on 23 known pathogenic somatic indels
