Fig. 1.
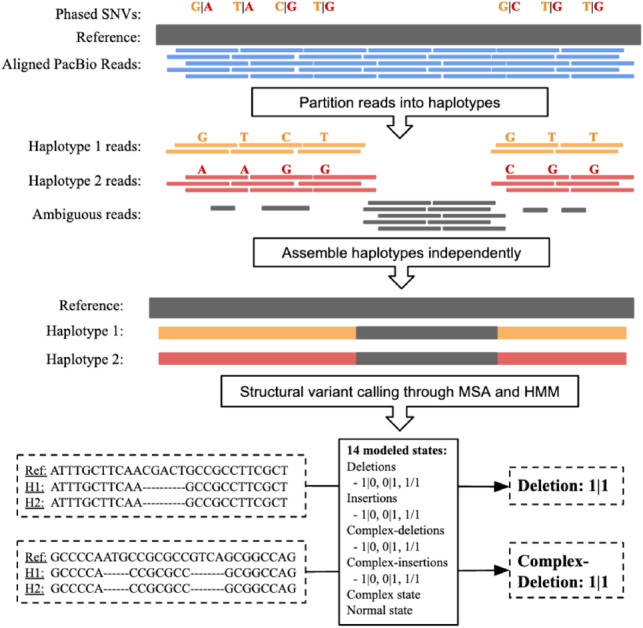
MsPAC workflow. Input reads are partitioned into haplotypes using phased SNVs. Next, partitioned reads are assembled independently. For each interval, the reference sequence and assembled contigs are multiple sequences aligned. Lastly, an HMM evaluates the MSA to return the final SV callset
