Fig. 2.
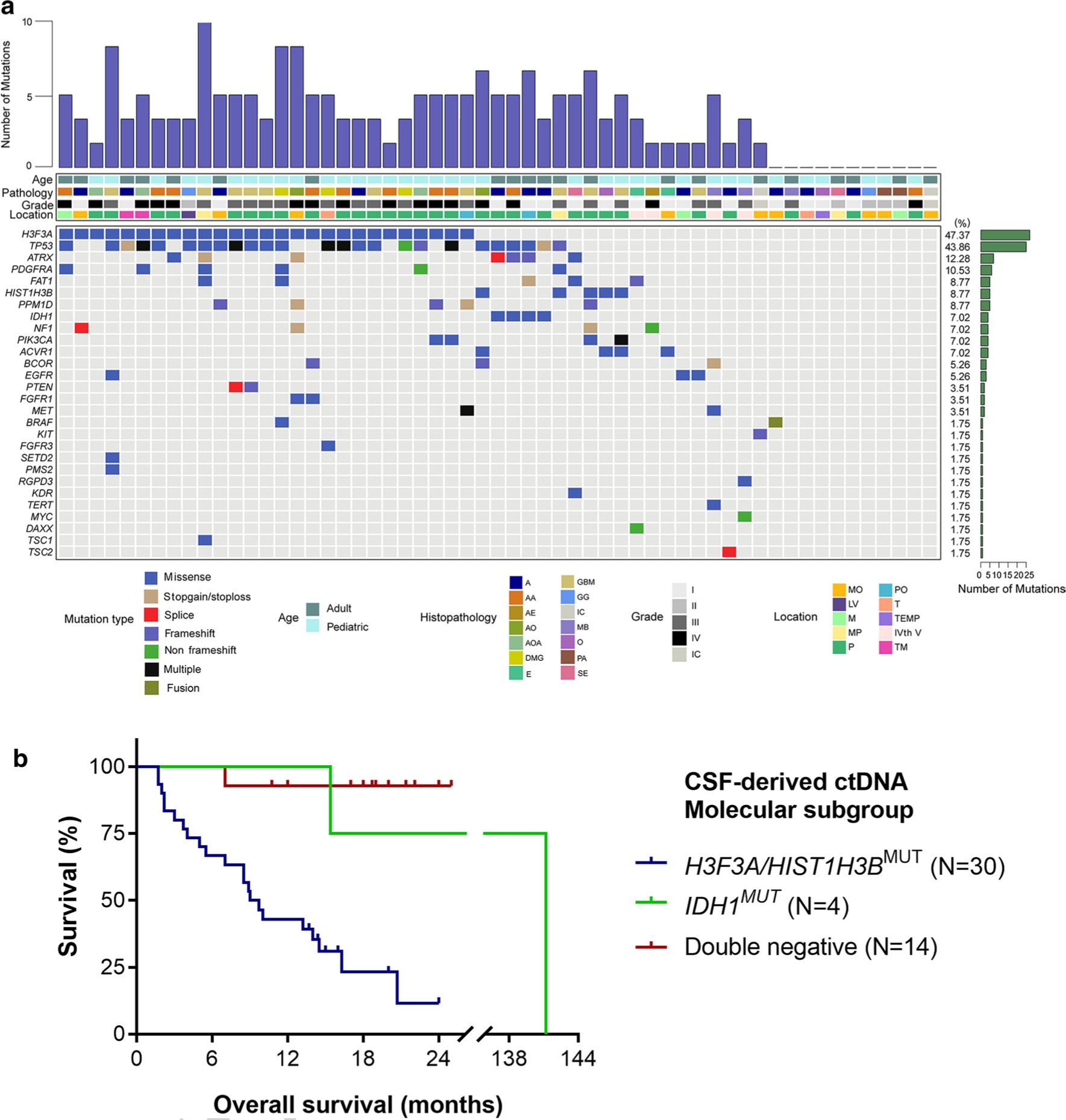
Mutational characteristics for cerebrospinal fluid cell-free DNA. a CSF ctDNA was successfully isolated from patients with brainstem tumors (N = 57). We used a panel-based next generation sequencing approach to identify recurrent tumor-specific alterations in the majority of CSF ctDNA samples. The most frequently mutated genes included H3F3A, TP53, ATRX and PDGFRA. Tumors were located in the following locations: MO medullary oblongata, P pontine, TM thalamus and midbrain, M midbrain, MP midbrain and pontine, PO pontine and medullary oblongata, TEMP temporal lobe, LV lateral ventricle, T thalamus, IVth V the fourth ventricle, IC inconclusive. The following histologies were included in our study: A astrocytoma AA anaplastic astrocytoma, DMG diffuse midline glioma, GBM glioblastoma, AOA anaplastic oligoastrocytoma, PA pilocytic astrocytoma, GG ganglioglioma, E ependymoma, AO anaplastic oligodendroglioma, SE subependymoma, O oligodendroglioma, AE anaplastic ependymoma, MB medulloblastoma. b Kaplan–Meier estimates of overall survival in brainstem and thalamic tumors based on molecular subgroups from CSF-derived ctDNA. The H3F3A/HIST1H3B mutant subgroup has a significantly worse survival of 9.35 months as compared to the IDH mutant group (141.2 months, p = 0.0318) and the double negative group (only one death in this subgroup, undefined median OS, p < 0.0001)
