Figure 4.
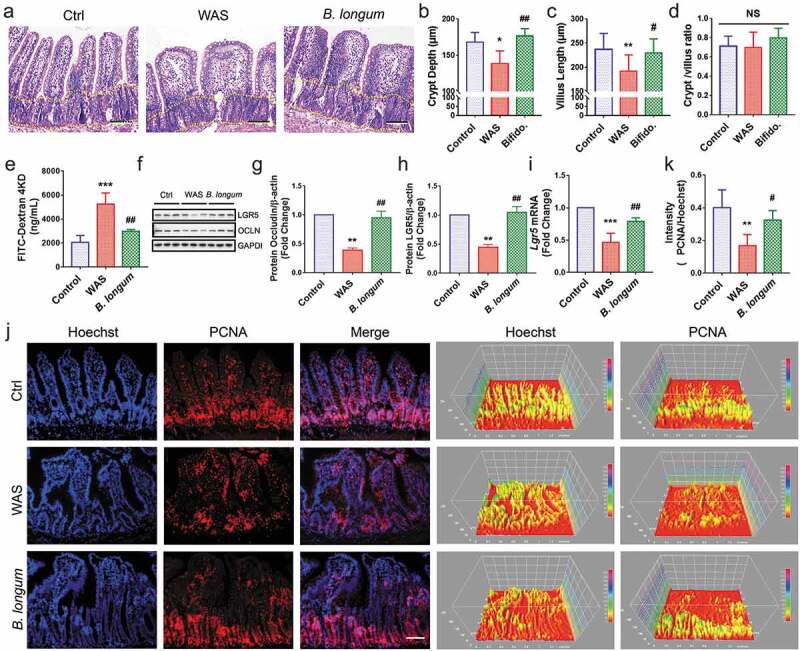
Bifidobacterium longum (B.longum) restored intestinal epithelial cell proliferation and enhanced mucosal repair in water avoidance stress (WAS) rats. (a) H&E staining of representative ileal sections of the rats from the three groups. The dashed line indicates the position of intestinal crypt. Scale bar = 100 μm. (b–d) Morphometric quantification of the intestinal crypt depth (b), villus height (c), and crypt/villus ratio (d) from five well-preserved crypts per slide of 8 rats from each group. Data are shown as the mean ± SD, (B) *, p = .0173 of Ctrl vs WAS, ##, p = .0014 of B. longum vs WAS (C).**, p = .0096 of Ctrl vs WAS, #, p = .0436 of B. longum vs WAS. (D) p = .9710 of Ctrl vs WAS, p = .2626 of B. longum vs WAS. (e) Intestinal permeability assay to detect intestinal permeability by measuring the concentration of FITC-D4000 in the rat serum from the three groups (n = 4 per group). Data are shown as the mean ± SD, ***, p = .0001 of Ctrl vs WAS, ##, p = .0018 of B. longum vs WAS. (f–h) Western blot analysis of occludin and LGR5 protein expression in the ileal tissue of 3 representative individual rats from each group. Quantification of GAPDH-normalized band intensity by Image-Pro Plus; data are shown as the mean ± SD, (g) **, p = .0023 of Ctrl vs WAS, ##, p = .0037 of B. longum vs WAS, n = 3, (h) **, p = .0022 of Ctrl vs WAS, ##, p = .0014 of B. longum vs WAS, (k) **, p = .0019 of Ctrl vs WAS, ##, p = .0271 of B. longum vs WAS, n = 3. (i) QPCR analysis of Lgr5 mRNA expression dissected from ileal tissues of the three groups. Data are shown as the mean ± SD, ***, p = .0007 of Ctrl vs WAS, ##, p = .0095 of B. longum vs WAS, n = 3. (j) Representative images of intestinal epithelial cells of ileal sections of rats from the three groups with PCNA (red) staining. The intensity of the cell nucleus and PCNA was quantified by Image J software. Scale bar = 100 μm. Results in (J) are shown as the average values of 5 sections per rat of 5 rats from each group. Data are shown as the mean ± SD, *, p = .0019 of Ctrl vs WAS, #, p = .0271 of B. longum vs WAS.
