Figure 3.
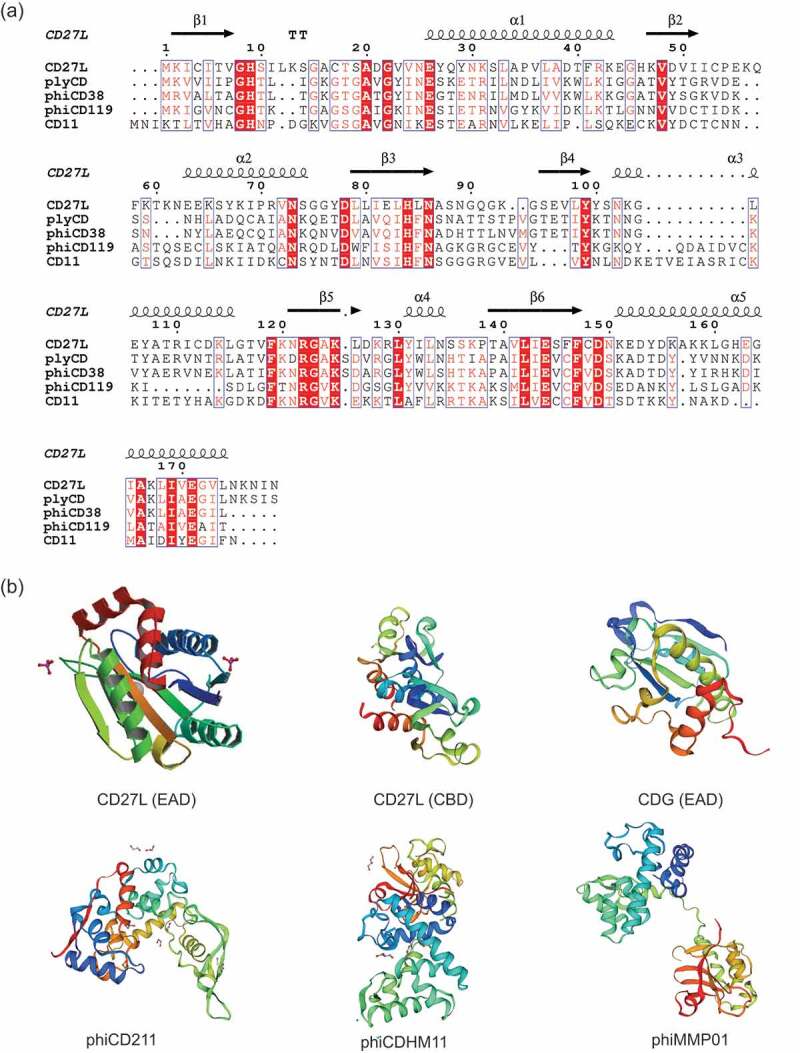
Sequence and structural analysis of C. difficile endolysins. (a) Sequence of the amidase containing catalytic domain of CD27L aligned to other amidase containing endolysins of C. difficile. Multiple-sequence alignment was performed by Clustal Omega and visualized by ESPript using CD27L endolysin structure (PDB code 3QAY) as the query. The position of amino acid residues based on CD27L amidase domain is shown. Secondary structure of CD27L1–179 is displayed, with arrows indicating beta strands and ribbons indicating alpha helices. Conserved residues can be visualized as white text on a red background, while amino acids with similar properties written in red text; (b) protein 3D structure of enzymatically active domains (EAD) and cell-wall binding domains (CBD). Homology modeling was performed by Swiss-model server (https://swissmodel.expasy.org/). The predicted model of CDG, phiCD211, phiCDHM11 and phiCDMMP01 were generated using protein data bank (PDB) templates 2L47, 5WQW, 4HPE and 4FDY, respectively.
