Figure 6.
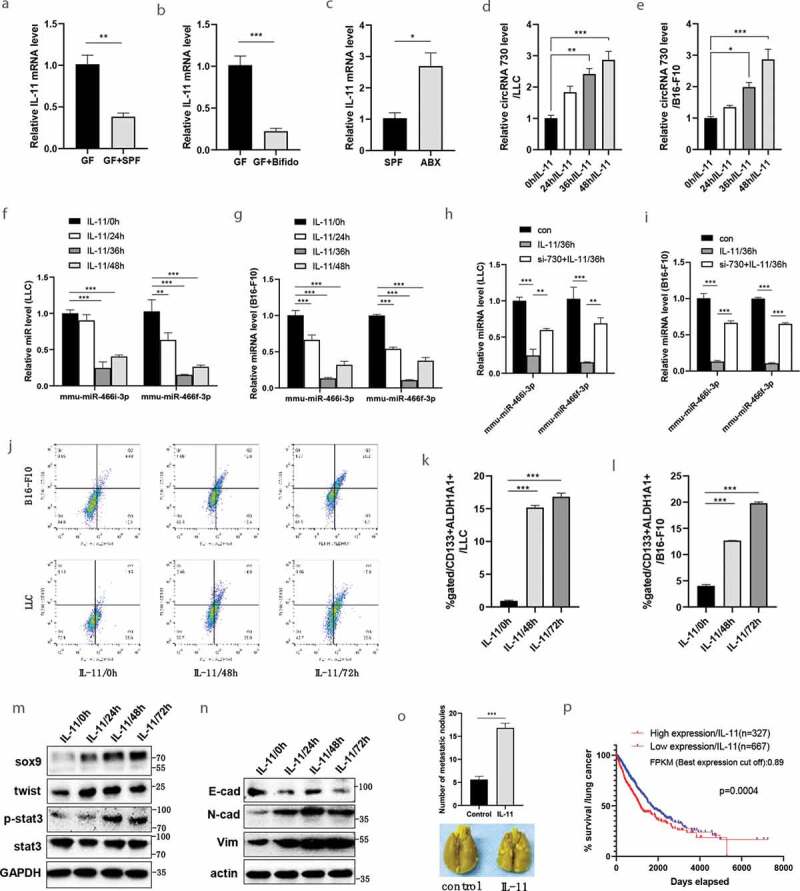
Gut microbiota regulates cancer metastasis through IL-11-mediated circRNA/microRNAs/sox9 axis. A-C, According to our RNA-seq results, transcript levels of IL-11 in tumor tissue were determined by qRT-PCR between GF and GF/SPF, GF and GF/Bifido, SPF and SPF/ABX groups. D-E, By LLC (d) or B16-F10 (e) cell line, to determine the effect of IL-11 on mmu_circ_0000730 expression by RT-qPCR. F-G, By LLC (f) or B16-F10 (g) cell line, to determine the effect of IL-11 on the expression of mmu-miR-466i-3p and mmu-miR-466 f-3p. H-I, IL-11 downregulates the expression of mmu-miR-466i-3p and mmu-miR-466 f-3p, while si-RNA-circ730 reverses these actions, by LLC (h) or B16-F10 (i) cell line. J-L, Regulatory role of IL-11 on cancer stem cells. CD133, ALDH1A1 alone or CD133/ALDH1A1 co-labeled cancer stem cells were examined by flow cytometry (J, K&L). M, IL-11 activates the STAT3 signaling pathway and promotes the expression of cancer stem cell marker genes such as SOX9 and twist in LLC cells. N, IL-11 promotes epithelial-mesenchymal transition (EMT) marker genes such as N-cadherin, vimentin and E-cadherin by western blot in LLC cells. O, IL-11 promotes tumor metastasis. Metastasis evaluations were performed in two sets and 6–8 mice per group, representative images (O, bottom). P, Survival analysis by TCGA data (https://www.proteinatlas.org) showed that high expression of IL-11 contributed to a reduced survival rate in lung cancer patients, P values are shown [log-rank (Mantel-Cox) analysis]. In Kaplan-Meyer plots by gene expression (IL-11), the thresholds of “high” and “low” were set according to FPKM (the best expression cut off). Data are shown as mean ± SEM from three experiments performed in triplicate (a-l). Statistical analysis: t test (A,B,C,O), one-way ANOVA (d,e), two-way ANOVA (F,G,H,I,K,L). *P < .05; **P < .01; ***P < .001.
