ABSTRACT
Type-I interferon (IFN-I) cytokines are produced by immune cells in response to microbial infections, cancer and autoimmune diseases, and subsequently, trigger cytoprotective and antiviral responses through the activation of IFN-I stimulated genes (ISGs). The ability of intestinal microbiota to modulate innate immune responses is well known, but the mechanisms underlying such responses remain elusive. Here we report that the intracellular sensors stimulator of IFN genes (STING) and mitochondrial antiviral signaling (MAVS) are essential for the production of IFN-I in response to lactic acid bacteria (LAB), common gut commensal bacteria with beneficial properties. Using human macrophage cells we show that LAB strains that potently activate the inflammatory transcription factor NF-κB are poor inducers of IFN-I and conversely, those triggering significant amounts of IFN-I fail to activate NF-κB. This IFN-I response is also observed in human primary macrophages, which modulate CD64 and CD40 upon challenge with IFN-I-inducing LAB. Mechanistically, IFN-I inducers interact more intimately with phagocytes as compared to NF-κB-inducers, and fail to activate IFN-I in the presence of phagocytosis inhibitors. These bacteria are then sensed intracellularly by the cytoplasmic sensors STING and, to a lesser extent, MAVS. Accordingly, macrophages deficient for STING showed dramatically reduced phosphorylation of TANK-binding kinase (TBK)-1 and IFN-I activation, which resulted in lower expression of ISGs. Our findings demonstrate a major role for intracellular sensing and STING in the production of IFN-I by beneficial bacteria and the existence of bacteria-specific immune signatures, which can be exploited to promote cytoprotective responses and prevent overreactive NF-κB-dependent inflammation in the gut.
KEYWORDS: Lactic acid bacteria, beneficial microbes, interferon, STING, MAVS
Introduction
Type-I IFN (IFN-I) such as IFN-α and IFN-β are polypeptides produced by innate immune cells in response to infectious agents, particularly viral pathogens1. IFNs are important to induce antimicrobial responses in an autocrine and paracrine manner via the synthesis of interferon-stimulated genes (ISGs), and to modulate innate immune responses that result in a balanced natural killer cell response and effective antigen presentation to activate immunological memory through specific T and B cells responses. IFN-I can also display protective roles against bacterial infections, cancer and autoimmune diseases.2 Innate immune cells produce IFN-I upon recognition of microbe-associated molecular patterns (MAMPs) via pattern recognition receptors (PRRs). The first described PRRs were Toll-like receptors (TLRs) and Nucleotide-binding oligomerization domain (NOD)-like intracellular receptors (NLRs). These PRRs activate an intracellular signaling cascade that converges on the pro-inflammatory transcription factor nuclear factor kappa-B (NF-κB), which is crucial in regulating anti-microbial responses in the mucosa.3 The production of IFN-I requires the additional and concomitant activation of IFN regulatory factors (IRFs), which can be achieved via the TLR adaptors myeloid differentiation primary response 88 (MyD88) and TIR domain-containing adapter-inducing IFN (TRIF).4 More recently, the cytosolic molecules stimulator of interferon genes (STING) and mitochondrial antiviral signaling (MAVS) have been identified as potent IFN-I inducers in response to nucleic acids.5 STING is an endoplasmic reticulum-localized transmembrane protein that becomes activated in the presence of cytosolic DNA and the production of 2‘,3‘-cyclic GMP-AMP (cGAMP) by the cGAMP synthase (cGAS).6–9 In addition, STING can also recognize other cyclic dinucleotides (CDN) of prokaryotic origin.6,10 MAVS is a mitochondria-localized transmembrane protein that is activated by retinoic acid inducible gene (RIG)-I and melanoma differentiation-associated protein (MDA)-5, two cytosolic sensors of 5‘ tri- and di-phosphate RNA and stretches of dsRNA.11–13 Both STING and MAVS coordinate the activation of TANK-binding kinase (TBK)-1 and IRF3, which translocates into the nucleus to initiate IFN production.14,15 The role of STING and MAVS in inducing protective immune responses in the presence of cytosolic DNA and RNA such as those from viral infections or intracellular pathogenic bacteria is well known,16,17 and in addition, STING has recently been shown to mediate sensing and autophagy of live gram-positive bacteria.18 However, the impact of STING and intracellular sensing on immune responses mediated by gut microbes remains relatively unexplored.
Humans live in symbiosis with beneficial microbes that inhabit different parts of the body, including the gastrointestinal and respiratory tracts.19,20 Beneficial microbes are composed of commensal bacteria that are critical to maintain homeostasis due to their involvement in appropriate host immune responses. One of the most significant bacterial groups associated with host beneficial properties are lactic acid bacteria (LAB),21 typical commensals that play an important role in activating epithelial cells, antigen-presenting cells and phagocytes.22 LAB interact with innate immune cells via numerous MAMPs including pili (fimbriae), peptidoglycans, lipo-teichoic acids, exopolysaccharides and many other components of the cell wall and membrane,22 that are recognized extracellularly by TLR and NLR and trigger NF-κB responses.21,22 A few studies have also reported that LAB are capable of inducing protective pro-inflammatory responses that depend not only on NF-κB activation, but also on IRFs.23,24 The specific mechanisms that LAB utilize to activate IRFs are unknown, but it has been reported that the production of type-I interferons (IFNs) requires the presence of TLRs in endosomal compartments, including TLR2 and TLR3.23,24
Here we demonstrate that different LAB species induce specific immune responses in human macrophages and primary cells, and that those that activate IFN-I responses fail to trigger NF-κB activation. Contrary to current dogma in which beneficial bacteria are mostly sensed by TLRs, we show that IFN-I-inducing LAB are sensed intracellularly via cytosolic nucleic acid sensors and that IFN-I production and ISG induction is dependent on STING and, to a lesser extent, MAVS. Our findings provide a novel perspective into the interactions between gut microbes and human cells that have implications in the use of and manipulation of beneficial commensal bacteria to modulate human immune responses.
Results
LAB activate NF-κB or IFN-I responses in THP-1 macrophages
The ability of 12 LAB strains (Table 1) to trigger the activation of the inflammatory transcription factors NF-κB and IRF-3 was evaluated in human differentiated THP-1 cells. We employed 2 lines of THP-1 monocytes expressing Gaussia luciferase (GLuc) under the control of either the NF-κB promoter or the promoter of the IRF3-dependent gene IFN-induced protein with tetratricopeptide repeats (IFIT)-1. The cells were differentiated for 48 h and subsequently exposed to live or dead LAB before GLuc activity was measured in the media and presented as a fold increase over non-stimulated macrophages (Figure 1(a)). The majority of LAB species tested – Streptococcus thermophilus, Pediococcus acidilactici, Lactobacillus sakei, Lactobacillus kunkeei, Lactobacillus casei, and Enterococcus faecalis - induced significant NF-κB activation and, with the exception of E. faecalis, this activation was enhanced in response to live bacteria compared to inactivated bacterial cells. By contrast, we found that these isolates were poor inducers of IFIT1 activation either dead or alive. Interestingly, two bacterial species – Pediococcus pentosaceus and Lactobacillus plantarum – were able to induce a significant IFIT1 activation. This IFIT1 response was only observed with viable bacteria and was especially high with L. plantarum. Interestingly, neither P. pentosaceus nor L. plantarum enhanced NF-κB activation when used as viable bacteria. To visualize this contrasting behavior, we then plotted the recorded NF-κB vs IFIT1 activation for each LAB species and drew arbitrary cutoffs for NF-κB activation (~20 fold) and IFIT1 (~10 fold) based on the most common response observed in all bacterial species tested (Figure 1(b)). As expected most LAB grouped in the top left quadrant (Q1) as powerful NF-κB agonists, but poor IFIT1 inducers. Strikingly, L. plantarum and P. pentosaceus appeared in the bottom right quadrant (Q4) as remarkably strong IFIT1 inducers, especially L. plantarum. The 2 L. lactis species occupied the bottom left quadrant (Q2) as intermediate NF-κB and IFIT1 inducers. We then performed dose-dependent exposures with L. plantarum and P. pentosaceus and showed that a challenge with 1 L. plantarum and 10 P. pentosaceus bacterial cells per macrophage was sufficient to trigger IFIT1 activation (Figure 1(c)). The highest IFIT1 response was observed with a ratio of 25 bacteria per macrophage. Our screening therefore indicated that some LAB could activate responses converging on IFN-I production.
Table 1.
Lactic Acid Bacteria (LAB) isolates used in this study
| Species | Strain | Internal code | Source | Reference | Donor/Culture collection |
|---|---|---|---|---|---|
| Enterococcus faecalis | NCTC 8213 | EF | Human babies’ faeces | 25 | Surrey Culture Collection |
| Lactobacillus casei | I-1518 | LC | Probiotic Drink | 26 | CNCM |
| Lactobacillus kunkeei | O29 | LK | Bee pollen | This study | |
| Lactobacillus plantarum | WCFS-1 | LP | Human saliva | 27 | Prof M Kleerebezem, Wageningen University, the Netherlands |
| Lactobacillus sakei | Lb790 | LS1 | Meat | 28 | Dr L Axelsson, Nofima AS, Norway |
| Lactobacillus sakei | L45 | LS2 | Norwegian fermented dried sausage | 29 | Prof I Ness, Norwegian University of Life Sciences |
| Lactococcus lactis | BBS4 | LL1 | Spanish fermented dried sausage | 30 | Prof PE Hernandez Cruza, Universidad Complutense de Madrid |
| Lactococcus lactis | IO-1 | LL2 | Kitchen sink water | 31 | Prof H Yoshikawa, Tokyo University of Agriculture |
| Pediococcus acidilactici | PAC1.0 | PA1 | Fermented food | 32 | Prof J Kok, Groningen University, the Netherlands |
| Pediococcus acidilactici | E1 | PA2 | Raw honey | This study | |
| Pediococcus pentosaceus | NCTC 990 | PP | Dried beer yeast | 33 | Surrey Culture Collection |
| Streptococcus thermophilus | I-1613 | ST | Yogurt | 34 | CNCM |
Figure 1.
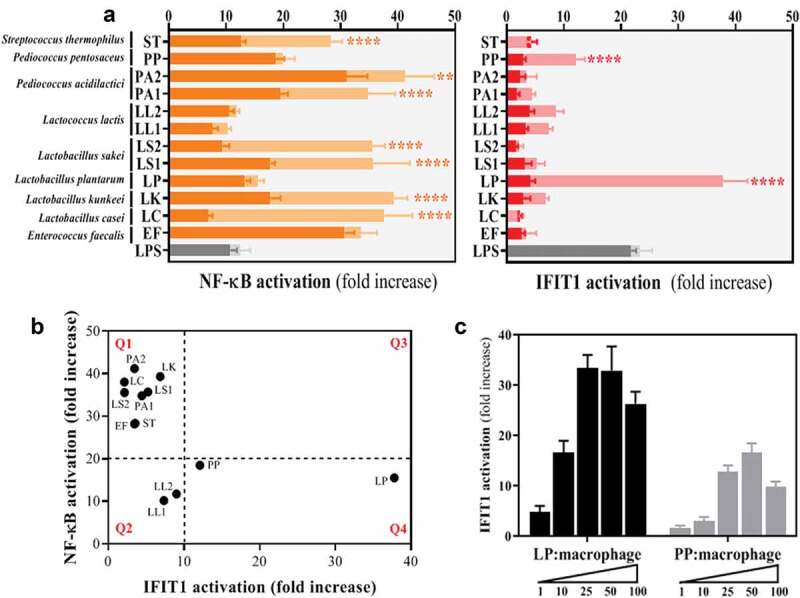
Analysis of 12 LAB strains reveals that viable cells of Lactobacillus plantarum (LP) and Pediococcus pentosaceus (PP) significantly induce IFIT1 activation in THP-1 macrophages in a dose-dependent manner. (a) Response of PMA-differentiated THP-1 macrophages to LAB isolates as a measurement of NF-κB (orange) and IFIT1 (red) activation. GLuc activation is presented as a fold increase over a non-stimulated condition. LAB were used at a ratio of 1 macrophage per 100 inactivated bacterial cells (dark orange and red) or 25 viable bacteria (light orange and red). LPS (gray bars) was used as a positive control in all screens. For each of the isolates, a comparative statistical analysis was carried out between inactivated and viable cells using the Student’s t-test (**p < .01; ****p < .001). (b) Correlation between NF-κB and IFIT1 activation in PMA-differentiated THP-1 macrophages exposed to LAB at a ratio of 1 macrophage per 25 viable bacteria. Activation is presented as a fold increase over a non-stimulated condition. Fold increases of 20 and 10 were selected as arbitrary thresholds for NF-κB and IFIT1 activation, respectively, to divide the graph into quadrants (Q1, Q2, Q3 and Q4). (c) IFIT1 activation in PMA-differentiated THP-1 macrophages challenged with viable cells of LP (black bars) and PP (gray bars) at increasing macrophage:bacteria ratios of 1:1, 1:10, 1:25, 1:50 and 1:100. The activation is presented as a fold increase over a non-stimulated condition.
L. plantarum and P. pentosaceus activate IFN-I production
To address expression of IFN-I and verify the activation observed using luciferase reporters, we used an IFN-stimulated responsive element (ISRE)-based bioassay and a commercial ELISA against the NF-κB-dependent inflammatory cytokine TNF-α to quantify the presence of TNF-α and IFN-I in the supernatants of macrophages exposed to each of the LAB isolates (Figure 2(a,b), respectively). Apart from L. plantarum, P. pentosaceus and isolates of L. lactis, the exposure of macrophages to all LAB isolates resulted in high amounts of TNF-α in the media, but very low levels of IFN-I. Conversely, L. plantarum and P. pentosaceus were capable of inducing a significant ISRE activation as indicative of effective production of IFN-I, especially in the case of L. plantarum (Figure 2(b)). Similarly to the luciferase-based results, when the production of TNF-α and IFN-I were plotted we identified a strong IFN-I signature for L. plantarum that correlated with a very low impact on TNF-α (bottom right quadrant Q4 of Figure 2(c)). The arbitrary cutoffs for production of TNF-α (~250 ng/mL) and IFN-I (~2 ISRE fold increase) were selected based on the most common response observed in all LAB tested. P. pentosaceus showed a more moderate IFN-I signature and a slightly higher TNF-α production than L. plantarum (next to the bottom left quadrant Q2 of Figure 2(c)), but this was still much lower than that observed with most of the LAB isolates (top left quadrant Q1 of Figure 2(c)). Taken together, our screening demonstrated that each LAB has the ability to stimulate different innate immune responses in human macrophages and that some are potent inducers of IFN-I.
Figure 2.
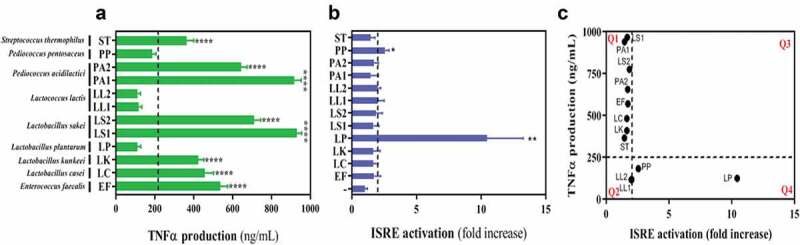
Lactobacillus plantarum (LP) and Pediococcus pentosaceus (PP) barely induce TNF-α production in THP-1 macrophages but significantly induce ISRE activation. (a-b) Supernatants obtained from THP-1 macrophages exposed to LAB isolates at a ratio of 1:25 were analysed for the presence of (a) TNF-α production using a commercial TNF-α ELISA and (b) ISRE activation using a pISRE-FLuc/RLuc bioassay. In both cases, isolates on the right-hand side of the dotted line induced statistically significant responses compared with the rest as determined by one way ANOVA followed by Fisher’s Least Significant Difference (LSD) Test (*p < .05; **p < .01; ****p < .001). (c) Correlation between TNF-α production and ISRE activation in THP-1 macrophages exposed to each of the LAB selected in this study at a ratio of 1:25. Doted lines indicate the thresholds used for TNF-α concentration (250 ng/mL) and ISRE activation (2-fold increase) to divide the graph into quadrants (Q1, Q2, Q3 and Q4).
L. plantarum and P. pentosaceus activate responses associated with IFN-I production in human PBMCs
Next we sought confirmation of LAB-induced IFN-I activation in human primary cells. We first employed an IFN-β ELISA to detect and quantify the presence of this cytokine in the supernatants of human monocyte-derived macrophages collected from PBMC and exposed to L. plantarum and P. pentosaceus for 2 h. We observed a peak of IFN-β production 8 h post-challenge that decreased at 12 h (Figure 3(a)). We then used flow cytometry to monitor the expression of the IFN-I-related markers CD64 and CD4035-37 in monocytes collected from PBMC from two healthy donors upon exposure to LAB. IFN-I expression has been shown to down-regulate CD6437 and upregulate CD40 in the presence of LAB.36 In agreement with this, we observed that only monocytes exposed to IFN-I-producing L. plantarum and P. pentosaceus showed a very significant reduction in CD64 expression (Figure 3(b)) and an increase in CD40 expression (Figure 3(c)). No significant changes were observed in the presence of L. casei, an isolate that showed a potent NF-κB response but poor IFN-I activation in our initial LAB screening. These findings therefore confirmed that LAB can trigger IFN-I responses in human primary immune cells.
Figure 3.
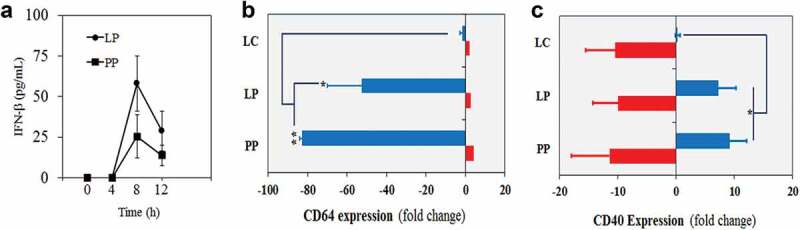
Lactobacillus plantarum (LP) and Pediococcus pentosaceus (PP) induce responses associated with IFN-I production in monocytes and macrophages from PBMCs. (a) Production of IFN-β in supernatants obtained from primary macrophages after 0, 4, 8 and 12 h of being exposed to LP and PP. The INF-β levels were calculated using the Human IFN-β Quantikine ELISA Kit. Data are representative of three healthy donors and are mean with SD from two biological replicates. (b-c) Expression of (b) CD64 and (c) CD40 in monocytes (blue) and neutrophils (red) exposed to LP and PP as well as L. casei (LC) as a control (no IFN-I inducer). The CD expression is presented as a percentage of fold change (increase or decrease) over a non-stimulated condition using unchallenged monocytes and neutrophils. Data are mean with SD from two healthy donors and the comparative analysis was carried out with two-way ANOVA and Tukey multiple comparison (*p < .05; **p < .01).
IFN-I-inducing LAB interact strongly with human monocytes and macrophages
We next determined the mechanism by which some LAB induce IFN-I production. First, we examined whether L. plantarum and P. pentosaceus interact with and/or are phagocytosed by human phagocytes (monocytes and macrophages). We carried out three independent assays using PBMCs, macrophages differentiated from PBMCs and THP1 macrophages, at a ratio of 1 phagocyte per 25 bacteria. This ratio ensures a maximum activation of IFIT1 in phagocytes exposed to either L. plantarum or P. pentosaceus (Figure 1(c)). Firstly, we incubated FITC-labeled LAB with human PBMC from healthy donors for 2 h and then processed the samples by flow cytometry. We identified three populations consisting of leukocytes, neutrophils and monocytes (Figure 4(a), top panels) and then assessed which of these had become positive for FITC (bottom panels). This assay showed that monocytes and neutrophils (blue and red populations) shifted significantly in the presence of L. plantarum and P. pentosaceus, but not with the NF-κB inducer L. casei. We quantitated these data and observed that contrary to L. casei, both L. plantarum and P. pentosaceus interacted significantly with monocytes (Figure 4(b)) and neutrophils (Figure 4(c)). Secondly, we differentiated PBMC monocytes into macrophages and observed them by confocal microscopy upon incubation with FITC-labeled L. plantarum and P. pentosaceus. We were able to visualize FITC+ cells (Figure 4(d)), indicating that a strong interaction occurred between LAB and primary human macrophages. Finally, we studied the effect of pharmacological inhibition of phagocytosis on the internalization of L. plantarum and P. pentosaceus in human THP-1 macrophages (Figure 5). We observed that the number of L. plantarum and P. pentosaceus that remain inside (and/or attached to) THP1 macrophages was significantly lower in the presence of the phagocytosis inhibitor cytochalasin D following a 2 h incubation in RPMI (Figure 5(a)). Concomitant with this decrease, the activation of the IFIT1-GLuc reporter upon exposure to L. plantarum and P. pentosaceus was significantly reduced in the presence of cytochalasin D (Figure 5(b)). Cytochalasin D did not affect bacterial viability (Figure 5(c)) nor affected reporter activation by unrelated agonists such as LPS (not shown). Taken together, these results strongly suggest that L. plantarum and P. pentosaceus interact and/or are internalized by human macrophages.
Figure 4.
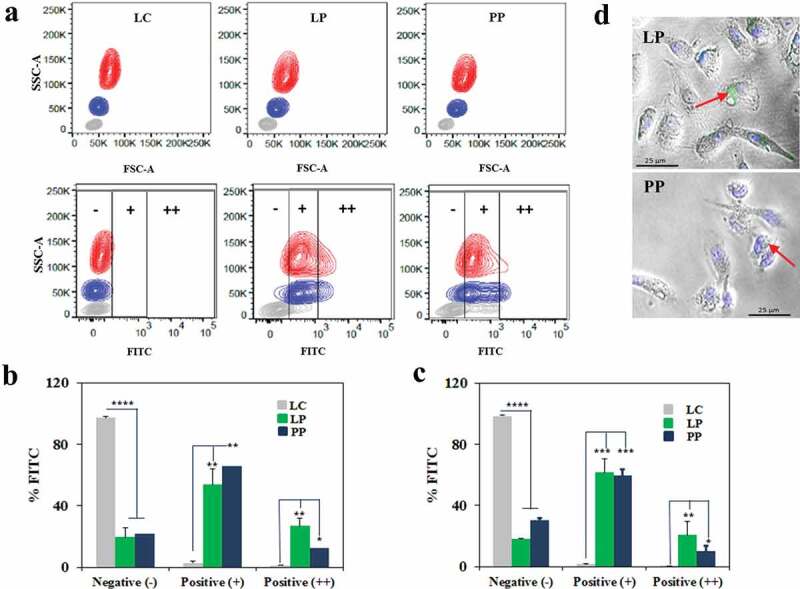
Human phagocytes from PBMCs interact with Lactobacillus plantarum (LP) and Pediococcus pentosaceus (PP). (a) Interaction of LP and PP as well as L. casei (LC) as a control with monocytes and neutrophils from PBMCs of two healthy donors. Viable bacterial cells were labeled with FITC and incubated in whole human blood for 1 h at a ratio of 1:25. Blood cell populations were distinguished based on side scatter area (SSC-A) versus forward scatter area (FSC-A), and separated into lymphocytes (gray), monocytes (blue) and neutrophils (red). Interaction with phagocytes was then observed in the FITC channel, where the intensity was divided into three subpopulations based on the positivity: no interaction (-), binding and/or uptake (+), and strong binding and/or uptake (++). (b-c) Percentage of (b) monocytes and (c) neutrophils in each of the three FITC subpopulations [no interaction (-), binding and/or uptake (+), and strong binding and/or uptake (++)] after exposure to LC (gray), LP (green) and PP (blue). The comparative analysis was carried out with two-way ANOVA and Tukey multiple comparisons (*p < .05; **p < .01; ***p < .005; ****p < .001). (d) Confocal microscopy images showing the detection of LP and PP at the vicinity and/or inside monocyte-derived macrophages (indicated with arrows). FITC and DAPI were used to label bacteria and the macrophage nucleus, respectively.
Figure 5.
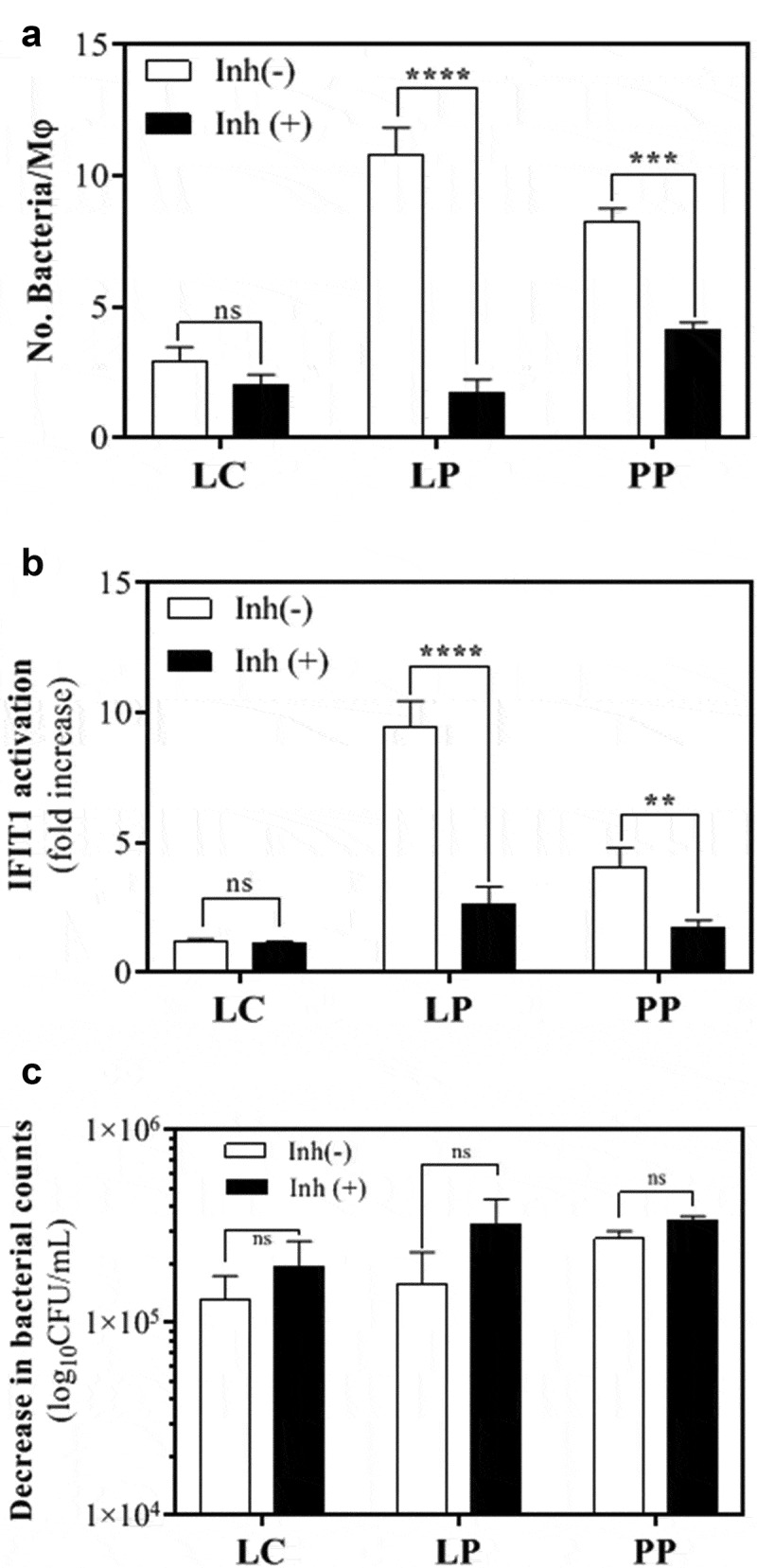
Human THP-1 macrophages interact with Lactobacillus plantarum (LP) and Pediococcus pentosaceus (PP). (a) Number of L. casei (LC), LP or PP cells that were internalized and/or attached to THP1 macrophages after a 2 h RPMI incubation in the absence (white) or presence (black) of the phagocytosis inhibitor cytochalasin D. The bacterial intake and/or attachment was calculated as the difference between the number of bacteria found in the supernatant of THP-1 cells at 0 and 2 h post-LAB challenge, and presented as number of bacteria per macrophage (Mφ). Comparative analysis was carried out using the Student t-test (ns, no significant; ***p < .005; ****p < .001). (b) IFIT1 activation in PMA-differentiated IFIT1-GLuc THP-1 macrophages exposed to LC, LP or PP in the absence (white) or presence (black) of cytochalasin D. The activation of IFIT1 is presented as a fold increase over a non-stimulated condition. The bacterial challenge was carried out at a ratio of 1 macrophage per 25 bacteria (**p < .01; ****p < .001; Student’s t-test). (c) Decrease in bacterial counts for LC, LP and PP after a 2 h RPMI incubation in the absence (white) or presence (black) of cytochalasin D. The bacterial decrease was calculated as the difference between the number of viable bacteria after 2 h of incubation and expressed as log10CFU/mL (ns, no significant; ***p < .005, ****p < .001; Student’s t-test).
STING and MAVS sense L. plantarum and P. pentosaceus to activate the IFN-I associated kinase TBK1
STING and MAVS are potent IFN-I inducers that respond to cytosolic MAMPs. Having observed that IFN-I-inducing LAB were internalized by macrophages, we explored whether these cytosolic sensors could account for the observed immune responses. We then used THP-1-IFIT1-GLuc cells deficient for STING (STING KO) or MAVS (MAVS KO).38 We differentiated these cells into macrophages and challenged them with viable cells of L. plantarum and P. pentosaceus before measuring GLuc activity. As expected, control cells responded strongly to L. plantarum and IFIT1 activation reached 10 and 40 fold increase at 12 and 24 h after challenge. However, this response was completely abrogated in STING KO cells (Figure 6(a)). The response in MAVS KO cells was also reduced, but this was less evident, being only statistically significant after 24 h of bacterial challenge. To verify the role of STING in L. plantarum-induced IFIT-1 activation, we examined the levels of phosphorylated TBK-1 in the lysates of cells exposed to different L. plantarum doses. Phosphorylated TBK-1 was absent in mock-challenged cells, but was readily detected 2 h after bacterial challenge in a dose-dependent manner (Figure 6(b)). In the absence of STING, we barely detected any phosphorylated TBK-1, whilst in the absence of MAVS, a reduction was appreciated in agreement with the reporter data. We then assessed the response to P. pentosaceus. As shown previously, exposure to P. pentosaceus triggered a lower response than that to L. plantarum and control cells showed elevated luciferase activity at 24 h post-challenge (Figure 6(c)). However, this was also drastically reduced in STING KO cells and partially reduced in the absence of MAVS (Figure 6(c)). Accordingly, levels of phosphorylated TBK1 were also diminished in the absence of STING or MAVS (Figure 6(d)). Collectively, our results demonstrate that L. plantarum and P. pentosaceus are sensed by mechanisms that involve the intracellular sensors STING and, to a lesser extent, MAVS.
Figure 6.
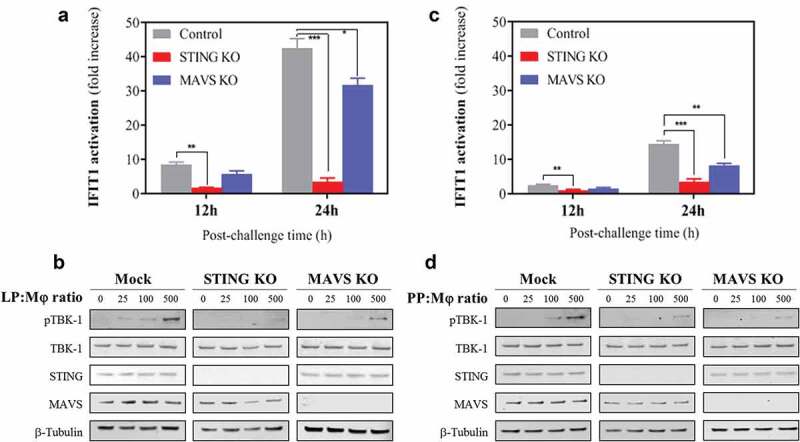
STING and MAVS sense Lactobacillus plantarum (LP) and Pediococcus pentosaceus (PP), resulting in the activation of the IFN-I associated kinase TBK-1. (a, c) IFIT1-GLuc activation in PMA-differentiated control (gray), STING knock-out (blue) and MAVS knock-out (red) THP-1 cells in response to (a) LP or (c) PP. IFIT1 activation is presented as a fold increase over a non-stimulated condition after 12 and 24 h of incubation using a ratio of 1 macrophage per 25 viable bacteria. For each time point, a comparative statistical analysis was carried out between the control and the knock-out cells using the Student’s t-test (*p < .05; **p < .01; ***p < .005). (b-d) Immunoblotting against the indicated proteins in whole-cell lysates from PMA-differentiated control, STING and MAVS knock-out THP-1 cells exposed to (b) LP or (d) PP at the indicated doses.
STING and MAVS are required to induce IFN-I in response to LAB
To evaluate the influence of STING and MAVS on the ability of L. plantarum and P. pentosaceus to activate IFN-I responses, we determined the induction of IFN-I and IFN-I-associated genes from STING and MAVS KO macrophages exposed to L. plantarum and P. pentosaceus at a ratio of 25 bacteria per macrophage (Figure 7). We first measured the relative expression of IFN-β, MxA and OAS1 in the challenged KO macrophages (Figure 7(a,b)). After 8 h of bacterial challenge with L. plantarum we detected a significant increase in the mRNA expression of IFN-β in the control cells (Figure 7(a)). In both STING and MAVS KO cells, this increase was significantly lower, in particular in the STING KO cells. Similarly, the absence of STING and MAVS in cells previously exposed to L. plantarum resulted in a significant decrease in the expression levels of MxA and OAS1, two well-known ISGs. The mRNA expression of these two ISGs, as well as IFN-β, also decreased in STING KO and MAVS KO cells challenged with P. pentosaceus as compared with the control cells (Figure 7(b)). However, only the reduction in IFN-β expression was found to be statistically significant. Finally, media from cells exposed to L. plantarum (the most potent inducer of IFN-I) were subjected to ISRE bioassay. In agreement with gene expression data, absence of STING and, to a lesser extent MAVS, significantly reduced the amount of biologically active IFN-I (Figure 7(c)). We observed similar results with KO cells exposed to P. pentosaceus but the reduction in functional IFN-I was less clear and equally significant from both KO cells (Figure 7(d)).
Figure 7.
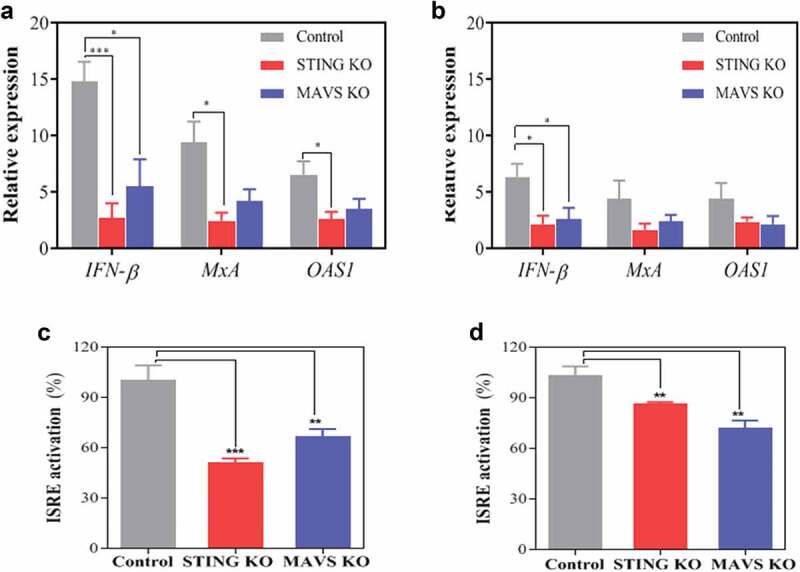
The IFN-I response of macrophages to Lactobacillus plantarum (LP) and Pediococcus pentosaceus (PP) is dependent on STING and MAVS. (a, b) Level of relative mRNA expression of IFN-β, MxA and OAS1 in IFIT1-GLuc THP-1 macrophages (control, gray) and their corresponding knockouts (KO) for STING (red) and MAVS (blue) after exposure to (a) LP or (b) PP for 8 h at a ratio of 1 macrophage per 25 viable bacteria. The relative expression of each gene is presented as a fold induction following normalization with the internal control gene TBP and nonstimulated THP-1 macrophages. A comparative statistical analysis was carried out between the control and the knockout cells using the Student’s t-test (*p < .05, ***p < .005). (C, D) ISRE activation from supernatants obtained from IFIT1-GLuc THP-1 macrophages (control, gray) and their corresponding knockouts (KO) for STING (red) and MAVS (blue) after exposure to (c) LP or (PP) for 24 h at a ratio of 1 macrophage per 25 viable bacteria. The ISRE activation for each cell line was calculated as a fold increase using a pISRE-FLuc/RLuc reporter cell line after 10 h of supernatant exposure and then indicated as a percentage relative to control. A comparative statistical analysis was carried out between the control and the knockout cells using the Student’s t-test (**p < .01; ***p < .005).
Discussion
Human cells express multiple PRRs aimed at detecting the presence of foreign molecules including those from bacteria and viruses. Such recognition activates signaling mechanisms that converge in the production of cytokines and chemokines. Currently, the central dogma is that commensal species are sensed by TLRs and NLRs and trigger activation of NF-κB. Certainly, LAB, a major commensal group, have been shown to activate NF-κB via TLR2,39 TLR940 and NLRs.41 A few studies have also shown that LAB are capable of inducing the production of IFN-I in innate immune cells,23,24,42,43 and when the mechanism has been reported, this has been linked with TLR2/3 recognition via endosomes.23,24 In this study, we report the production of IFN-I by LAB in human macrophages and demonstrate that such production is mediated by intracellular sensing driven by the nucleic acid sensors STING and MAVS. Our findings were generated from an initial-unbiased screen using different representative LAB species. This screen showed that macrophage challenge with LAB species such as E. faecalis, L. casei, L. sakei and P. acidilactici resulted in very high NF-κB activation, as widely reported in previous publications.44–47 However, unlike most of the selected LAB, L. plantarum and P. pentosaceus failed to significantly activate the NF-κB pathway, but were instead able to induce production of IFN-I in human macrophage-like cells and human primary phagocytes isolated from PBMCs. LAB induction of IFN-I production was species-specific since it was not observed for other lactobacilli and pediococci tested in our screen. This bacterial specificity has been confirmed with other strains of L. plantarum and P. pediococcus that we have recently isolated from animals.47,48 The level of IFN-I produced by these bacteria in macrophages was detectable by ELISA, but more importantly, it induced upregulation of CD40 and downregulation of CD64, both of which are well-known IFN-I-responsive markers.35–37 In this respect, Weiss et al. found that LAB that induce IFN-β activation are also able to stimulate CD40 in murine bone marrow derived dendritic cells,36 a positive convergence that we have confirmed here using human primary monocyte-derived macrophages. Furthermore, IFN-I triggered the production of ISGs in the form of MxA and OAS1, effectively activating an antiviral response. Our work therefore may provide explanations for how the composition of microbial communities and specific bacterial groups modulate innate immune responses during virus infection,49 an emerging field with impact on gastrointestinal and non-gastrointestinal infection as well as vaccines.
The IFN-I activation that we have recorded here required live bacteria and their interaction with paghocytes, as previously reported with dentritic cells stimulated with other LAB species.23 Alive cells of L. plantarum and P. penstosaceus triggered an endogenous IFN-I production that was significantly higher than that observed with inactivated cells. In general, macrophages exposed to heat-killed (or inactivated) bacterial cells activate TRL/NOD-like receptors-dependent pathways, whereas live bacteria activate other pathways that require phagocytosis and bacterial degradation in the phagolysosome, which may lead to the release of bacterial intracellular components into the cytosol.50 Recently, it has been shown that STING is essential to activate an integrated stress response to live Gram-positive bacteria particularly Listeria, and that part of this response involves the late production of IFN-I.18 STING activation occurred upon engagement with the CDN c-di-AMP secreted by these bacteria and therefore it bypassed cGAS.18,51,52 Whilst many clinically relevant Gram-positive bacteria including L. monocytogenes or Mycobacterium tuberculosis produce c-di-AMP and are hence susceptible to STING responses, it is unclear whether this is also the case for LAB. The synthesis of CDNs has been described in LAB, but very little is known about their role in bacterial physiology and host innate immune responses.53 More recent work has identified the oxidoreductase RECON as a high-affinity cytosolic sensor of CDNs including c-di-AMP,54 and it has been proposed that bacteria such as L. monocytogenes secrete c-di-AMP to inhibit RECON and relieve RECON-mediated NF-κB inhibition to promote bacterial spread.55 Therefore, if LAB were inducing IFN-I via c-di-AMP secretion, a concomitant increase in NF-κB response would have been expected. Our findings indicating that IFN-I-inducing LAB hardly activated NF-κB suggest that other PAMPs and mechanisms may be responsible for STING sensing. A possible explanation to account for the observed STING-dependent responses would be the release of bacterial DNA into the cytosolic millieu acting as an intracellular PAMP. We believe that the requirement for live LAB cells could be associated to the process of internalization and subsequent DNA recognition by professional phagocytes.
Recent studies have reported that the cytosolic sensors STING and MAVS are important to maintain gut homeostasis and prevent autoimmune conditions such as inflammatory bowel disease (IBD).56,57 Our results are in line with these latest discoveries. Although multiple commensals might be involved in beneficial nucleic acid responses in the gut, our study has identified L. plantarum as a potential key player. We have proved that L. plantarum interacts intimately with human phagocytes and that this close interaction is essential to activate IFN-I responses via STING and MAVS. We have also observed that L. plantarum failed to activate NF-κB, a transcription factor that drives pathology in many inflammatory disorders. In these, microbial dysbiosis and the continuous production of potent NF-κB-mediated inflammatory cytokines such as TNF-α drive perpetual inflammation. IFN-I is emerging as an important cytoprotective signature in mucosal tissues including the gut.58,59 Therefore, L. plantarum and their mechanisms could assist in targeting the underlying microbe-immune cell interactions and inducing beneficial anti-inflammatory cytokines. L. plantarum is a frequent inhabitant of mucosal tissues where a variety of immune cells including macrophages and dendritic cells lie and it is widely recognized as a potential immunomodulator. At present, it is unclear how L. plantarum is recognized by macrophage receptors and what properties this bacterium has to account for its ability to activate STING and MAVS. Our results comparing phagocytic uptake of L. plantarum with other LAB such as L. casei (Figure 4) indicates that the level of interaction with human phagocytes might be a prominent differential feature, which eventually leads to phagocytic uptake and intracellular processing of the bacterial cells. Although multiple factors may exist, one possibility explaining this enhanced interacting capacity is the ability of L. plantarum to express adhesins that facilitate interactions with other cells.60 The oral administration of L. plantarum as a probiotic is common and remarkably it has resulted in a significant reduction in respiratory tract infections in newborns.61 These probiotic features and the mechanisms underlying the IFN-I activation that we report in this work indicate that L. plantarum might be an excellent candidate to mitigate inflammation as well as enhancing protective antiviral responses.
In summary, our findings highlight the value of LAB and their molecules as potential mediators of host protective IFN-I responses via the cytosolic sensors STING and MAVS. A better understanding of the STING-mediated IFN-I regulation by L. plantarum and other LAB that have a very low impact on NF-κB-mediated pro-inflammatory cytokines could be helpful in the design of future probiotic therapies addressing human gut disorders.
Materials and methods
Ethics statement
All procedures with human blood samples have been approved by the Surrey University Ethics Committee in accordance with the Institutional Policy on the Donation and Use of Human Specimens in Teaching and Research and the national guidelines under which the institution operates. The blood protocol approval was under IRAS number 236477 and the sample storage was carried out under HTA license 12365.
Human peripheral blood mononuclear cells (PBMC) and macrophages
Whole fresh blood was collected in heparinized tubes from healthy individuals to carry out the phagocytosis assay. To generate macrophages, monocytes were first isolated from buffy coats of the healthy blood donors by Lymphoprep (Axis-Shield) density gradient centrifugation followed by plastic adherence in Roswell Park Memorial Institute (RPMI) 1640 (Life Technologies) supplemented with 15% fetal bovine serum (FCS, Seralab) and 1% Penicillin/Streptomycin (Pen/Strep, Life Technologies) at 37°C in an atmosphere of 5% CO2. The collected peripheral blood mononuclear cells (PBMCs) were incubated for 2 h to remove nonadherent cells by washing with PBS. The adherent monocytes were then detached using 10 mM EDTA in PBS at room temperature and plated for macrophage maturation in RPMI with FCS and Pen/Strep for 7 days.
THP-1 cell lines and macrophage differentiation
THP-1 cells were propagated in RPMI supplemented with 15% FCS and 1% Pen/Strep. THP-1-κB-GLuc (Lucia™ NF-κB) express and secrete Gaussia luciferase (GLuc) under the control of a synthetic 5x-κB promoter (Invivogen). THP-1-IFIT1-GLuc express and secrete Gluc under the control of the promoter of the IRF-3-dependent gene IFIT1 and were a gift from Veit Hornung.62 THP-1-IFIT1 deficient for STING or MAVS, and its corresponding control, were kindly provided by Greg Towers and have been previously described.38 THP-1 monocytes were differentiated into macrophages in RPMI supplemented with 20 ng/mL of phorbol 12-myristate 13-acetate (PMA) for 48 h as previously described.63 After the 48 h incubation medium was replaced for RPMI containing 2% FCS and 1% Pen/Strep and macrophages exposed to LAB. PMA was from Santa Cruz Biotechnology and kept in DMSO at 10 mg/mL.
Growth and maintenance of LAB
The LAB selected for this study is shown in Table 1 and are reference strains with accessible peer-reviewed publications and/or genome sequences with the exception of Pedioccocus acidilactici (isolate E1) and Lactobacillus kunkeei (isolate O29). The genomes of the newly-characterized isolates E1 and O29 and their corresponding sequencing reads, assemblies and metadata have been uploaded onto Genbank in BioProject PRJNA544274. All the isolates were grown in MRS broth (Oxoid) at 37ºC without any aeration for 24 h, and maintained as −80°C frozen stocks in their appropriate media with the addition of 15% glycerol.
Bacterial challenge
PMA-differentiated THP-1-NF-κB-GLuc and THP-1-IFIT1-GLuc macrophages were exposed to inactivated and viable LAB cells. The inactivated cells were used as heat-treated (70°C, 2 h) LAB pellets at a ratio of 100 cells per 1 macrophage and incubated for 24 h, while viable cells were incubated simultaneously with the macrophages for 2 h in RPMI without Pen/Strep at different doses as indicated in the figure legends before being replaced for fresh RPMI with antibiotics to incubate for a further 22 h. To measure Gluc activity, media were collected from the macrophage cultures and transferred to white-bottom 96-well plates, which were read in a Clariostar plate reader (BMG Biotech) in the presence of 2 μg/mL of coelenterazine (NanoLight Technology). LPS (Sigma Aldrich) was used at 0.2 mg/ml as a control for the activation of NF-κB and IFIT1. Activation of NF-κB or IFIT1 was calculated as a fold increase ± SD over the measurements recorded for unchallenged macrophages.
Bacterial interaction with THP-1 macrophages
In order to estimate the number of LAB that interact with and/or are internalized by THP-1 macrophages, we carried out an extracellular survival assay as previously described64 with minor modifications. Macrophages were incubated with viable LAB at a ratio of 25 bacteria per phagocyte in RPMI containing 2% FCS for 2 h, at 37°C and 5% CO2. Macrophages were treated with the phagocytosis inhibitor cytochalasin D at a final concentration of 10 μM (Sigma-Aldrich) and subsequently challenged with LAB. LAB were also incubated without macrophages under the same experimental conditions (with or without cytochalasin D). Supernatants were then collected at time points 0 and 2 h to calculate and compare the number of bacteria that remained viable and detached from THP-1 macrophages. Bacterial enumeration was performed by quantitative plating of serial dilutions on MRS agar plates.
Phagocyte assay with human peripheral blood mononuclear cells (PBMC)
Approximately 106 leukocytes were mixed 1:1 with 10 mM EDTA in PBS and challenged with LAB that were previously labeled with the FITC dye (Sigma), at a ratio of 25 bacteria per cell. The phagocytosis incubation was then carried out at 37°C in an orbital shaker for 1 h and subsequently treated with 1x RBC lysis solution (Biolegend) following incubation at room temperature for 15 min. The cells were washed twice with 10nM EDTA in PBS, resuspended in PBS and analyzed on FACS Celesta instrument (BD Biosciences). Flow cytometry analysis based on forward (FSC) and side (SSC) scatter was used to distinguish the main blood cell populations based on their size and granularity (lymphocytes vs. phagocytes), while the FITC channel was used to measure bacterial interaction with blood cells. The resulting SCC/FITC plots were then used to quantify the LAB intake by phagocytes.
CD expression in phagocytes from human PBMCs
In order to monitor the expression of CD40 and CD64 in monocytes and neutrophils exposed to LAB we carried out the phagocytosis assay as described above but with some modifications. Leukocytes and bacteria were mixed at 1:100 ratios and incubated for 1 h. After phagocytosis incubation antibodies against human CD40-BV510 and CD64-PE/Dazzle (Biolegend), or the corresponding isotype controls, were added to the samples and incubated for 20 min at room temperature. The fixable viability dye Zombie NIR (Biolegend) was also used for a live/dead stain as per manufacturer’s instructions. Cells were then fixed with RBC lysis buffer and the CD expression compared to the matching isotype control and no-bacteria controls to calculate expression levels in each of the blood cells populations identified by FACS.
Microscopy studies on monocyte-derived macrophages from human PBMCs
Monocyte-derived macrophages were challenged for 2 h with FITC-labeled LAB as described above for THP-1 cells. After phagocytosis macrophages were fixed for 20 min with 1% paraformaldehyde (PFA), washed with PBS without Calcium or Magnesium and stained with DAPI for imaging on an EVOS fluorescent microscopy.
SDS-PAGE and immunoblotting
LAB-challenged THP-1 cells were lysed in radio-immuno-precipitation assay (RIPA) buffer supplemented with protease and phosphatase inhibitors (Roche) and benzonase at 250 U/ml (Sigma). Lysates were rotated for 30 min at 4°C and subsequently denatured for 5 min at 95°C in the presence of loading buffer and resolved by SDS-PAGE. The samples were then transferred to nitrocellulose membranes (GE Healthcare) using a Trans-Blot semidry transfer unit (Bio-Rad). Membranes were blocked in 0.1% Tween/phosphate-buffered saline supplemented with 5%-skimmed milk (Sigma) and subjected to immunoblotting with the following primary antibodies at the indicated dilutions: phosphorylated TBK-1 Ser17 (Abcam; 1:5,000), TBK−1 (Abcam; 1:5,000) and α-tubulin (Upstate Biotech; 1:10,000). Primary antibodies were detected using IRDye-conjugated secondary antibodies in an Odyssey infrared imager (LI-COR Biosciences). Images were analyzed using Odyssey software.
Quantitative RT-PCR
PMA-differentiated THP−1 cells were exposed to LAB for 8 h and cells were collected for RNA extraction using the High Pure RNA Isolation Kit (Roche Diagnostics Limited) following manufacturer’s instructions. RNA was reverse transcribed using the SuperScript ® III First-Strand Synthesis System from Invitrogen and the resulting cDNA diluted 1:5 in water. cDNA was then used as a template for real-time amplification on a QuantStudio 7 Flex Real-Time PCR system (Applied Biosystems) using SYBR Green Master Mix (Applied Biosystems) and specific primers for: TATA box Binding Protein (TBP) (forward, 5ʹTGCACAGGAGCCAAGAGTGAA; reverse, 5ʹ CACATCACAGCTCCCCACCA), myxovirus resistance gene A (MxA) (forward, 5ʹ CCCCAGTAATGTGGACATCG; reverse, 5ʹ ACCTTGTCTTCAGTTCCTTTGT), human 2ʹ5’-oligoadenylate synthetase 1 (OAS1) (forward, 5ʹ TGTGTGTGTCCAAGGTGGTA; reverse, 5ʹ TGATCCTGAAAAGTGGTGAGAG), and human IFNβ (described previously65) using three technical replicates of samples obtained from three biological replicates. Expression of each gene was normalized to the house-keeping gene TBP, and these values were then normalized to the non-stimulated control cells to yield a fold induction.
ISRE-based IFN-I bioassay
Cell culture supernatants from LAB–challenged, PMA-differentiated THP-1 macrophages were collected after 24 h of incubation and transferred to 96-well plates containing HEK293T cells previously transfected for 24 h with 70 ng/well of a reporter plasmid expressing firefly luciferase (FLuc) under the control of ISRE and 10 ng/well of a control plasmid expressing Renilla luciferase (RLuc) (Promega) using polyethylenimine (PEI; Sigma) at a ratio of 1:2 (μg of DNA:μl of PEI). After 10 h the cells were lysed in passive lysis buffer (Promega) and the FLuc and RLuc activities measured in the Clariostar plate reader (BMG Biotech). The FLuc/RLuc ratios were then calculated for each well and normalized to mock-infected THP-1 samples to be presented as a fold increase of ISRE activation.
ELISA cytokine detection
Human TNF-α production was detected and quantified in the supernatants of LAB-challenged PMA-differentiated THP-1 cells using the eBioscience Human TNF-α ELISA Ready-SET-Go kit as indicated by the manufacturer’s instructions. The Human IFN-β Quantikine ELISA Kit (R&D systems) was used to detect and quantify IFN-β in supernatants from primary macrophages exposed to LAB for 2 h. Supernatants were collected every 4 h for 12 h after the exposure to LAB.
Statistical analysis
Statistical analysis was performed using GraphPad Prism. Data are presented as means ± standard deviation (SD) and are representative of one experiment of at least three independent experiments. Data from experiments with human peripheral blood mononuclear cells are representative of two or three healthy donors and are mean with SD from two biological replicates. Statistical significance between one sample and its corresponding control was determined using the Student’s t-test and within a group of samples using one way ANOVA followed by Fisher’s Least Significant Difference (LSD) Test.
Acknowledgments
We thank Veit Hornung (University of Munich, Germany) and Greg Towers (University College London, UK) for providing reagents. We would also like to thank Sophie Brooks and Tiago Marques Pedro for their technical assistance.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Funding Statement
This study was funded by internal start-up funds from the University of Surrey (to JGM) and BBSRC grant BB/M003647/1 (to CMdM). TC holds a University of Surrey School of Biosciences and Medicine PhD studentship.
Author Contributions
JGM planned and performed experiments, carried out data analysis and prepared and edited the manuscript. BI performed experiments, aided in data analysis, and aided in preparing and editing the manuscript. TC conducted technical experiments and edited the manuscript. FME designed experiments and aided in preparing and editing the manuscript. CMdM designed and performed experiments, aided in data analysis, and prepared and edited the manuscript. All authors read and approved the final manuscript.
Disclosure of Potential Conflicts of Interest
The authors have no conflicts of interest to declare.
Statement of Ethics
All procedures with human blood samples have been approved by the Surrey University Ethics Committee in accordance with the Institutional Policy on the Donation and Use of Human Specimens in Teaching and Research and the national guidelines under which the institution operates. The blood protocol approval was under IRAS number 236477 and the sample storage was carried out under HTA licence 12365.
References
- 1.Ivashkiv LB, Donlin LT.. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trinchieri G. Type I interferon: friend or foe? J Exp Med. 2010;207:2053–2063. doi: 10.1084/jem.20101664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pasparakis M. Role of NF-kappaB in epithelial biology. Immunol Rev. 2012;246:346–358. doi: 10.1111/j.1600-065X.2012.01109.x. [DOI] [PubMed] [Google Scholar]
- 4.Radoshevich L, Dussurget O. Cytosolic innate immune sensing and signaling upon infection. Front Microbiol. 2016;7:313. doi: 10.3389/fmicb.2016.00313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishikawa H, Barber GN. The STING pathway and regulation of innate immune signaling in response to DNA pathogens. Cell Mol Life Sci. 2011;68:1157–1165. doi: 10.1007/s00018-010-0605-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, Hopfner KP, Ludwig J, Hornung V. cGAS produces a 2 ‘-5 ‘-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498:380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun LJ, Wu JX, Chen ZJ. Cyclic GMP-AMP Synthase (cGAS) is a cytosolic DNA sensor that activates type I interferon pathway by generating a second messenger. J Immunol. 2013;190:786–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao TS, Fitzgerald KA. The cGAS-STING pathway for DNA sensing. Mol Cell. 2013;51:135–139. doi: 10.1016/j.molcel.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sauer JD, Sotelo-Troha K, von Moltke J, Monroe KM, Rae CS, Brubaker SW, Hyodo M, Hayakawa Y, Woodward JJ, Portnoy DA, et al. The N-Ethyl-N-nitrosourea-induced goldenticket mouse mutant reveals an essential function of sting in the in vivo interferon response to listeria monocytogenes and cyclic dinucleotides. Infect Immun. 2011;79:688–694. doi: 10.1128/IAI.00999-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seth RB, Sun LJ, Ea CK, Chen ZJJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappa B and IRF3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 12.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S.. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. [DOI] [PubMed] [Google Scholar]
- 13.Zeng WW, Sun LJ, Jiang XM, Chen X, Hou FJ, Adhikari A, Xu M, Chen ZJ.. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T.. IKK epsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4:491–496. [DOI] [PubMed] [Google Scholar]
- 15.Sharma S, tenOever BR, Grandvaux N, Zhou GP, Lin RT, Hiscott J. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;300:1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- 16.Buss C, Opitz B, Hocke AC, Lippmann J, van Laak V, Hippenstiel S, Krüll M, Suttorp N, Eitel J.. Essential role of mitochondrial antiviral signaling, IFN regulatory factor (IRF)3, and IRF7 in Chlamydophila pneumoniae-mediated IFN-beta response and control of bacterial replication in human endothelial cells. J Immunol. 2010;184:3072–3078. [DOI] [PubMed] [Google Scholar]
- 17.Schmolke M, Patel JR, de Castro E, Sanchez-Aparicio MT, Uccellini MB, Miller JC, Manicassamy B, Satoh T, Kawai T, Akira S, et al. RIG-I detects mRNA of intracellular Salmonella enterica serovar Typhimurium during bacterial infection. Mbio. 2014;5:e01006–14. doi: 10.1128/mBio.01006-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moretti J, Roy S, Bozec D, Martinez J, Chapman JR, Ueberheide B, Lamming DW, Chen ZJ, Horng T, Yeretssian G, et al. STING senses microbial viability to orchestrate stress-mediated autophagy of the endoplasmic reticulum. Cell. 2017;171:809–23 e13. doi: 10.1016/j.cell.2017.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brestoff JR, Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat Immunol. 2013;14:676–684. doi: 10.1038/ni.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martens K, Pugin B, De Boeck I, Spacova I, Steelant B, Seys SF, Lebeer S, Hellings PW.. Probiotics for the airways: potential to improve epithelial and immune homeostasis. Allergy. 2018;73:1954–1963. [DOI] [PubMed] [Google Scholar]
- 21.Pessione E. Lactic acid bacteria contribution to gut microbiota complexity: lights and shadows. Front Cell Infect Microbiol. 2012;2:86. doi: 10.3389/fcimb.2012.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hevia A, Delgado S, Sanchez B, Margolles A. Molecular players involved in the interaction between beneficial bacteria and the immune system. Front Microbiol. 2015;6:1285. doi: 10.3389/fmicb.2015.01285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weiss G, Maaetoft-Udsen K, Stifter SA, Hertzog P, Goriely S, Thomsen AR, Paludan SR, Frokiaer H. MyD88 drives the IFN-beta response to Lactobacillus acidophilus in dendritic cells through a mechanism involving IRF1, IRF3, and IRF7. J Immunol. 2012;189:2860–2868. [DOI] [PubMed] [Google Scholar]
- 24.Kawashima T, Kosaka A, Yan H, Guo Z, Uchiyama R, Fukui R, Kaneko D, Kumagai Y, You D-J, Carreras J, et al. Double-stranded RNA of intestinal commensal but not pathogenic bacteria triggers production of protective interferon-beta. Immunity. 2013;38:1187–1197. doi: 10.1016/j.immuni.2013.02.024. [DOI] [PubMed] [Google Scholar]
- 25.Farrow JA, Jones D, Phillips BA, Collins MD. Taxonomic studies on some group D streptococci. J Gen Microbiol. 1983;129:1423–1432. doi: 10.1099/00221287-129-5-1423. [DOI] [PubMed] [Google Scholar]
- 26.Oozeer R, Leplingard A, Mater DDG, Mogenet A, Michelin R, Seksek I, Marteau P, Dore J, Bresson J-L, Corthier G, et al. Survival of Lactobacillus casei in the human digestive tract after consumption of fermented milk. Appl Environ Microb. 2006;72:5615–5617. doi: 10.1128/AEM.00722-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kleerebezem M, Boekhorst J, van Kranenburg R, Molenaar D, Kuipers OP, Leer R, Tarchini R, Peters SA, Sandbrink HM, Fiers MWEJ, et al. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc Natl Acad Sci U S A. 2003;100:1990–1995. doi: 10.1073/pnas.0337704100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schillinger U, Lucke FK. Antibacterial activity of Lactobacillus sake isolated from meat. Appl Environ Microbiol. 1989;55:1901–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mortvedt CI, Nissen-Meyer J, Sletten K, Nes IF. Purification and amino acid sequence of lactocin S, a bacteriocin produced by Lactobacillus sake L45. Appl Environ Microbiol. 1991;57:1829–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodriguez JM, Cintas LM, Casaus P, Horn N, Dodd HM, Hernandez PE, Gasson MJ.. Isolation of nisin-producing Lactococcus lactis strains from dry fermented sausages. J Appl Bacteriol. 1995;78:109–115. [DOI] [PubMed] [Google Scholar]
- 31.Kato H, Shiwa Y, Oshima K, Machii M, Araya-Kojima T, Zendo T, Shimizu-Kadota M, Hattori M, Sonomoto K, Yoshikawa H, et al. Complete genome sequence of Lactococcus lactis IO-1, a lactic acid bacterium that utilizes xylose and produces high levels of L-lactic acid. J Bacteriol. 2012;194:2102–2103. doi: 10.1128/JB.00074-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gonzalez CF, Kunka BS. Plasmid-associated bacteriocin production and sucrose fermentation in pediococcus acidilactici. Appl Environ Microbiol. 1987;53:2534–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skerman VBD, McGowan V, Sneath PHA. Approved lists of bacterial names. Med J Aust. 1980;2:3–4. [DOI] [PubMed] [Google Scholar]
- 34.McNulty NP, Yatsunenko T, Hsiao A, Faith JJ, Muegge BD, Goodman AL, Henrissat B, Oozeer R, Cools-Portier S, Gobert G, et al. the impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci Transl Med. 2011;3:106ra106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bauvois B, Nguyen J, Tang R, Billard C, Kolb JP. Types I and II interferons upregulate the costimulatory CD80 molecule in monocytes via interferon regulatory factor-1. Biochem Pharmacol. 2009;78:514–522. doi: 10.1016/j.bcp.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 36.Weiss G, Christensen HR, Zeuthen LH, Vogensen FK, Jakobsen M, Frokiaer H. Lactobacilli and bifidobacteria induce differential interferon-beta profiles in dendritic cells. Cytokine. 2011;56:520–530. doi: 10.1016/j.cyto.2011.07.024. [DOI] [PubMed] [Google Scholar]
- 37.Van Weyenbergh J, Lipinski P, Abadie A, Chabas D, Blank U, Liblau R, Wietzerbin J.. Antagonistic action of IFN-beta and IFN-gamma on high affinity Fc gamma receptor expression in healthy controls and multiple sclerosis patients. J Immunol. 1998;161:1568–1574. [PubMed] [Google Scholar]
- 38.Tie CH, Fernandes L, Conde L, Robbez-Masson L, Sumner RP, Peacock T, Rodriguez‐Plata MT, Mickute G, Gifford R, Towers GJ, et al. KAP1 regulates endogenous retroviruses in adult human cells and contributes to innate immune control. EMBO Rep. 2018;19:e45000. doi: 10.15252/embr.201745000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karrasch T, Kim JS, Muhlbauer M, Magness ST, Jobin C. Gnotobiotic IL-10-/-;NF-kappa B(EGFP) mice reveal the critical role of TLR/NF-kappa B signaling in commensal bacteria-induced colitis. J Immunol. 2007;178:6522–6532. doi: 10.4049/jimmunol.178.10.6522. [DOI] [PubMed] [Google Scholar]
- 40.Hiramatsu Y, Satho T, Irie K, Shiimura S, Okuno T, Sharmin T, Uyeda S, Fukumitsu Y, Nakashima Y, Miake F, et al. Differences in TLR9-dependent inhibitory effects of H2O2-induced IL-8 secretion and NF-kappa B/I kappa B-alpha system activation by genomic DNA from five Lactobacillus species. Microbes Infect. 2013;15:96–104. doi: 10.1016/j.micinf.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 41.Zeuthen LH, Fink LN, Frokiaer H. Toll-like receptor 2 and nucleotide-binding oligomerization domain-2 play divergent roles in the recognition of gut-derived lactobacilli and bifidobacteria in dendritic cells. Immunology. 2008;124:489–502. doi: 10.1111/imm.2008.124.issue-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pereyra BS, Falcoff R, Falcoff E, Lemonnier D. Interferon induction by Lactobacillus bulgaricus and Streptococcus thermophilus in mice. Eur Cytokine Netw. 1991;2:299–303. [PubMed] [Google Scholar]
- 43.Kitazawa H, Tomioka Y, Matsumura K, Aso H, Mizugaki M, Itoh T, Yamaguchi T.. Expression of mRNA encoding IFN alpha in macrophages stimulated with Lactobacillus gasseri. FEMS Microbiol Lett. 1994;120:315–321. [DOI] [PubMed] [Google Scholar]
- 44.Kim YG, Ohta T, Takahashi T, Kushiro A, Nomoto K, Yokokura T, Okada N, Danbara H. et al. Probiotic Lactobacillus casei activates innate immunity via NF-kappa B and p38 MAP kinase signaling pathways. Microbes Infect. 2006;8:994–1005. [DOI] [PubMed] [Google Scholar]
- 45.Zou J, Shankar N. Roles of TLR/MyD88/MAPK/NF-kappaB signaling pathways in the regulation of phagocytosis and proinflammatory cytokine expression in response to E. faecalis infection. PLoS One. 2015;10:e0136947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jung JY, Shin JS, Lee SG, Rhee YK, Cho CW, Hong HD, Lee KT.. Lactobacillus sakei K040706 evokes immunostimulatory effects on macrophages through TLR 2-mediated activation. Int Immunopharmacol. 2015;28:88–96. [DOI] [PubMed] [Google Scholar]
- 47.Stedman A, Maluquer de Motes C, Lesellier S, Dalley D, Chambers M, Gutierrez-Merino J. Lactic acid Bacteria isolated from European badgers (Meles meles) reduce the viability and survival of Bacillus Calmette-Guerin (BCG) vaccine and influence the immune response to BCG in a human macrophage model. BMC Microbiol. 2018;18:74. doi: 10.1186/s12866-018-1210-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bravo M, Combes T, Martinez-Estrada F, Cerrato R, Rey J, Garcia-Jimenez W, Fernandez-Llario P, Risco D, Gutierrez-Merino J.. Lactobacilli isolated from wild boar (Sus scrofa) antagonize Mycobacterium bovis Bacille Calmette-Guerin (BCG) in a species-dependent manner Front. Microbiol. 2019;10:1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mazel-Sanchez B, Yildiz S, Schmolke M. Menage a trois: virus, host, and microbiota in experimental infection models. Trends Microbiol. 2019;27:440–452. doi: 10.1016/j.tim.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 50.Charrel-Dennis M, Latz E, Halmen KA, Trieu-Cuot P, Fitzgerald KA, Kasper DL, Golenback DT.. TLR-independent type I interferon induction in response to an extracellular bacterial pathogen via intracellular recognition of its DNA. Cell Host Microbe. 2008;4:543–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Woodward JJ, Iavarone AT, Portnoy DA. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science. 2010;328:1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Burdette DL, Vance RE. STING and the innate immune response to nucleic acids in the cytosol. Nat Immunol. 2013;14:19–26. doi: 10.1038/ni.2491. [DOI] [PubMed] [Google Scholar]
- 53.Papadimitriou K, Alegria A, Bron PA, de Angelis M, Gobbetti M, Kleerebezem M, Lemos JA, Linares DM, Ross P, Stanton C, et al. Stress physiology of lactic acid bacteria. Microbiol Mol Biol Rev. 2016;80:837–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McFarland AP, Luo S, Ahmed-Qadri F, Zuck M, Thayer EF, Goo YA, Hybiske K, Tong L, Woodward JJ.. Sensing of bacterial cyclic dinucleotides by the oxidoreductase RECON promotes NF-kappaB activation and shapes a proinflammatory antibacterial state. Immunity. 2017;46:433–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McFarland AP, Burke TP, Carletti AA, Glover RC, Tabakh H, Welch MD, Woodward JJ. et al. RECON-dependent inflammation in hepatocytes enhances listeria monocytogenes cell-to-cell spread. MBio. 2018;9:e00526–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li XD, Chiu YH, Ismail AS, Behrendt CL, Wight-Carter M, Hooper LV, Chen ZJ.. Mitochondrial antiviral signaling protein (MAVS) monitors commensal bacteria and induces an immune response that prevents experimental colitis. P Natl Acad Sci USA. 2011;108:17390–17395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Negishi H, Miki S, Sarashina H, Taguchi-Atarashi N, Nakajima A, Matsuki K, Endo N, Yanai H, Nishio J, Honda K, et al. Essential contribution of IRF3 to intestinal homeostasis and microbiota-mediated Tslp gene induction. Proc Natl Acad Sci U S A. 2012;109:21016–21021. doi: 10.1073/pnas.1219482110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kotredes KP, Thomas B, Gamero AM. The protective role of type I interferons in the gastrointestinal tract. Front Immunol. 2017;8:410. doi: 10.3389/fimmu.2017.00410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Uhlig HH, Powrie F. Translating immunology into therapeutic concepts for inflammatory bowel disease. Annu Rev Immunol. 2018;36:755–781. doi: 10.1146/annurev-immunol-042617-053055. [DOI] [PubMed] [Google Scholar]
- 60.Pretzer G, Snel J, Molenaar D, Wiersma A, Bron PA, Lambert J, de Vos WM, van der Meer R, Smits MA, Kleerebezem M, et al. Biodiversity-based identification and functional characterization of the mannose-specific adhesin of Lactobacillus plantarum. J Bacteriol. 2005;187:6128–6136. doi: 10.1128/JB.187.17.6128-6136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Panigrahi P, Parida S, Nanda NC, Satpathy R, Pradhan L, Chandel DS, Baccaglini L, Mohapatra A, Mohapatra SS, Misra PR, et al. A randomized synbiotic trial to prevent sepsis among infants in rural India. Nature. 2017;548:407–412. doi: 10.1038/nature23480. [DOI] [PubMed] [Google Scholar]
- 62.Mankan AK, Schmidt T, Chauhan D, Goldeck M, Honing K, Gaidt M, Kubarenko AV, Andreeva L, Hopfner K-P, Hornung V, et al. Cytosolic RNA: DNA hybrids activate the cGAS-STING axis. Embo J. 2014;33:2937–2946. doi: 10.15252/embj.201488726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Georgana I, Sumner RP, Towers GJ, Maluquer de Motes C.. Virulent poxviruses inhibit DNA sensing by preventing STING activation. J Virol. 2018;92:e02145–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ribes S, Ebert S, Regen T, Agarwal A, Tauber SC, Czesnik D, Spreer A, Bunkowski S, Eiffert H, Hanisch U-K, et al. Toll-like receptor stimulation enhances phagocytosis and intracellular killing of nonencapsulated and encapsulated Streptococcus pneumoniae by murine microglia. Infect Immun. 2010;78:865–871. doi: 10.1128/IAI.01110-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Odon V, Georgana I, Holley J, Morata J, Maluquer de Motes C. Novel class of viral ankyrin proteins targeting the host E3 ubiquitin ligase cullin-2. J Virol. 2018;92:e01374–18. [DOI] [PMC free article] [PubMed] [Google Scholar]


