Figure 1.
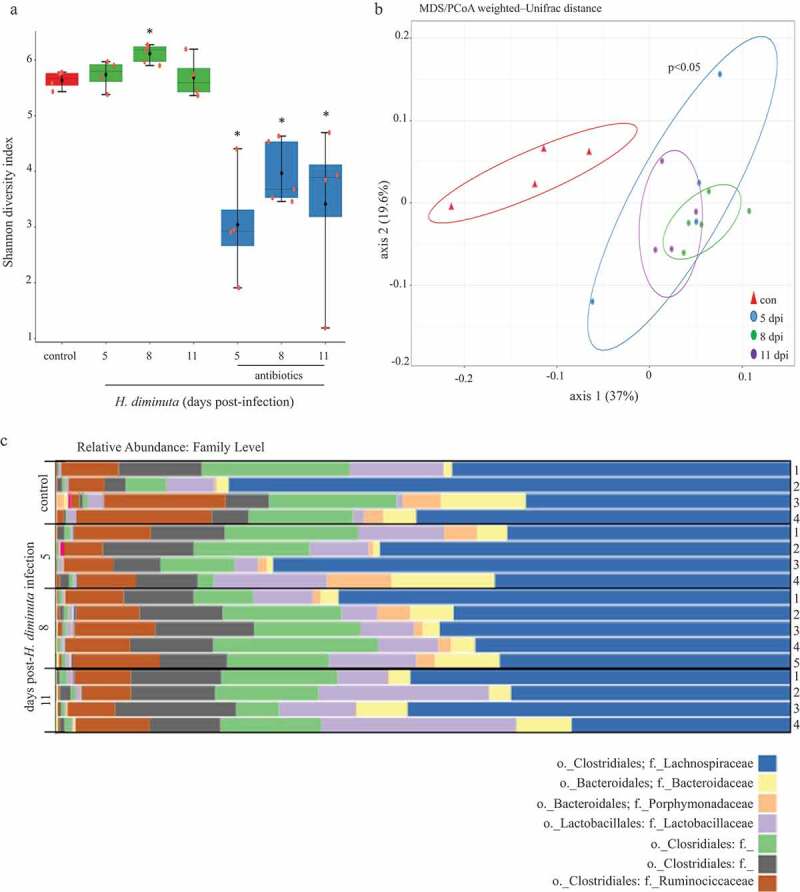
Sequence analysis of the V3-V4 region of the bacterial 16S rRNA gene reveals subtle shifts in the composition of colonic mucosa-associated bacteria following infection with H. diminuta. Male BALB/c mice were infected with 5 H. diminuta and assessed 5, 8 and 11 days post-infection (dpi). (A) Shannon diversity index (α-diversity) (horizontal line, median; black diamond, mean; box plots, 25–75% quartiles; vertical line, minimum and maximum value; * and **, p < .05 and p < .01 compared to control (uninfected). (B) Weighted-Unifrac distance metric for β-diversity, followed by a PERMANOVA test for significance, demonstrates that infection with H. diminuta changes the taxonomic composition of the colonic microbiota. (C) Relative family abundance based on amplicon sequence variants assessment. (D) Differential abundance analysis using DESeq2 comparing control and H. diminuta-infected mice (n = 4–5; * indicated Clostridium cluster XIVa across the time-line). (E) Bacterial colony-forming units (CFU) after 24 h of 37°C anaerobic culture of feces from control and 8-d-infected mice on M2GSC medium agar (n = 7–8 mice from two experiments; *, p < .05; o, order; f, family; g, genus)).
