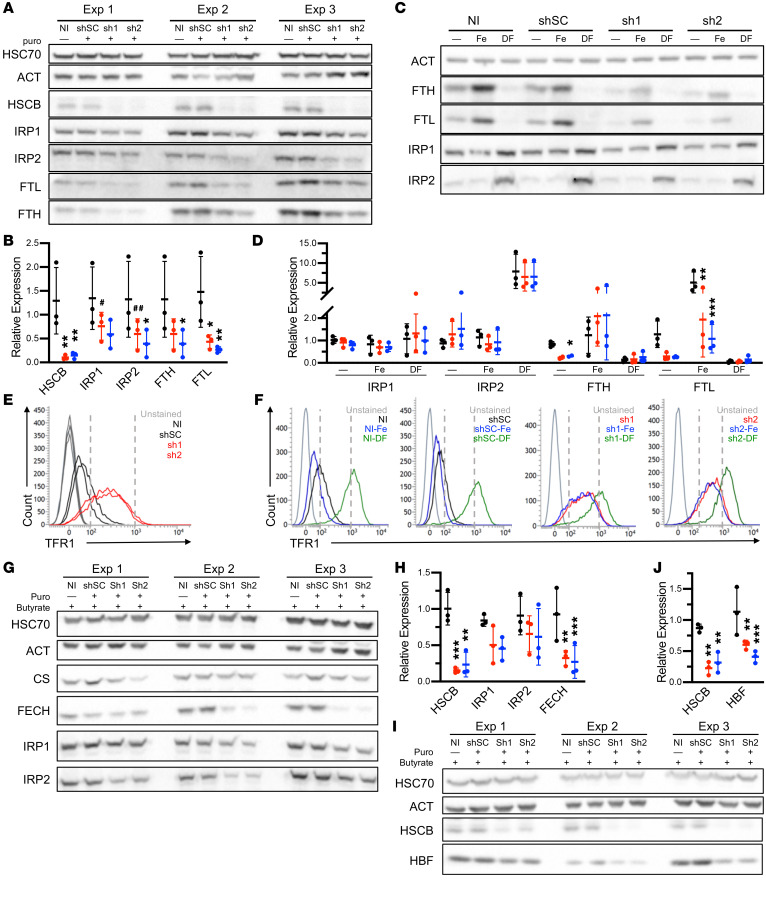
Figure 3. HSCB deficiency affects iron metabolism and hemoglobinization in K562 cells.
(A) Western blots of 3 independent experiments of iron metabolism proteins in uninduced K562 cells infected with viruses encoding scrambled (shSC) and HSCB-specific shRNAs (sh1 and sh2) for 10 days. (B) Quantification of Western blots in A. shSC, sh1, and sh2 are indicated by black, red, and blue, respectively. (C and D) Western blots (C) and quantification (D) of iron metabolism proteins in uninduced K562 cells treated with shRNAs as in A and either iron (Fe) or the iron chelator desferrioxamine (DF), demonstrating reduced expression but retained response of iron-regulated proteins to iron concentrations. One representative blot of 3 independent experiments is shown. Two ferritin heavy chain (FTH) outliers were removed based on Grubbs’s test P < 0.05. (E and F) Transferrin receptor 1 (TFR1) cell surface expression by flow cytometry of cells in A and C, respectively. (G–J) K562 cells treated with shRNAs as in A and differentiated by exposure to sodium butyrate for 96 hours. Western blotting demonstrates alterations in iron metabolism proteins (G and H) and impaired hemoglobin F expression (I and J). Three independent experiments are shown. For all panels, mean expression ± SD is normalized to noninfected (NI) cells. ANOVA, *P < 0.05, **P < 0.01, ***P < 0.001, #P = 0.06, ##P = 0.07 vs. shSC.