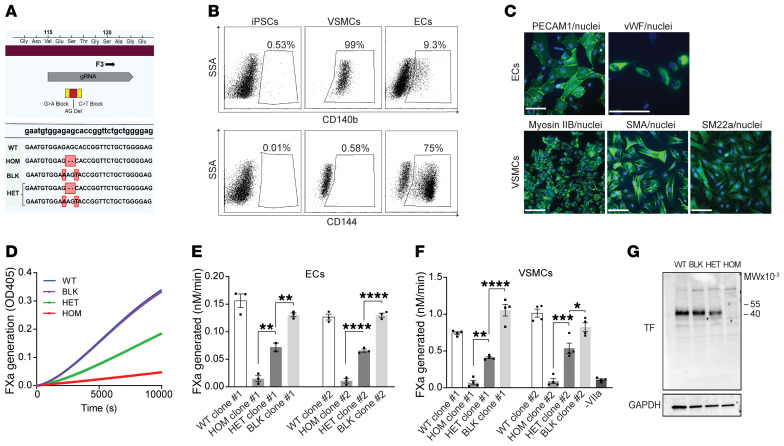
Figure 3. iPSCs heterozygous for the truncating F3 allele differentiated into vascular cells exhibit haploinsufficiency of coagulation initiation.
(A) CRISPR was used to engineer iPSCs heterozygous (HET) or homozygous (HOM) for the patient’s deletion (red). Synonymous blocking mutations (yellow) prevented editing of the second allele. WT (WT) and blocking mutant (BLK) iPSC lines provided additional controls. Edited bases (red boxes) and the site of gRNA homology (gray) are highlighted. (B) Representative fluorescence activated cell sorting plots demonstrating differentiation of WT iPSCs (left) into VSMCs (middle) and ECs (right) by staining for CD140b (upper panel) and CD144 (lower panel). (C) Validation of endothelial cell differentiation by indirect immunofluorescence staining for PECAM1 and von Willebrand factor (VWF); VSMC differentiation was validated using myosin IIb, smooth muscle actin (SMA), and smooth muscle protein 22-α (SM22a). Nuclei are stained with Hoechst. Scale bars: 100 µm. (D) Engineered iPSCs differentiated into ECs were evaluated for their ability to support the factor VIIa catalyzed conversion of factor X to factor Xa (FXa), as determined by cleavage of a chromogenic substrate. A representative kinetic course reflects mean ± SEM for 3 replicates. (E) Quantification of D; n = 3 independent experiments for 2 independently isolated iPSC clones differentiated into ECs (n = 2 for a single clone), 1-way analysis of variance. (F) iPSC clones differentiated into VSMCs were tested for FXa generation as in D and E, n = 4. (G) TF expression in differentiated VSMCs was determined by immunoblot with anti-TF and anti-GAPDH antibodies. See complete unedited blots in the supplemental material. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.