Abstract
Streptococcus oralis (S. oralis) has been recognized as a fatal pathogen to cause multiorgan failure by contributing to the formation of microthrombus. Coagulation and fibrinolysis systems have been found under the control of circadian clock genes. This study aimed to explore the correlation between BMAL1 and coagulation factor biosynthesis in S. oralis infection. Mice were administered S. oralis to induce sepsis, and HepG2 cells were also infected by S. oralis. The expression of BMAL1 of hepatocytes was downregulated in the S. oralis infection group, leading to the downregulation of coagulation factor VII (FVII) and the upregulation of the coagulation factor XII (FXII) in vitro and in vivo. Furthermore, we confirmed that the deficiency of BAML1 contributed to the elevation of FVII and the decline in FXII by constructing BMAL1-deficiency (Bmal1−/−) mice. The current result showed that BMAL1 regulates FVII directly. Thus, a novel insight into the coagulation abnormality in S. oralis infection was gained that may optimize the treatment of sepsis by rescuing the expression of BMAL1 in the liver.
Keywords: S. oralis, bmal1, FVII, FXII, coagulation factor biosynthesis
Introduction
Streptococcus oralis (S. oralis) is an oral biofilm-deriving opportunistic pathogen (Li et al., 2004) that has been found in endocarditis (Turnier et al., 2009) and cerebral abscess (Thiagarajan et al., 2016). Reportedly, in neutropenic patients, S. oralis induces sepsis (Penack et al., 2014; Shelburne et al., 2014) and inflicts death (Cornely and Schirmacher, 2001). Thus, the mechanisms of S. oralis-oriented abnormal coagulation to reduce the mortality need to be deduced. The overwhelming host response can progress to disseminated intravascular coagulation (DIC) by upregulating the activation of the coagulation cascade (Fourrier et al., 1992). It is characterized by excessive activated coagulation systems and suppressed fibrinolytic systems and is known to contribute to the formation of microthrombus, which leads to multiorgan failure (Vervloet et al., 1998). In this process, several coagulation factors patriciate in both classic clotting pathways (Amaral et al., 2004). In addition, most of the coagulation factors in coagulation cascade are generated by hepatocytes. Blood coagulation factor VII (FVII) and factor XII (FXII) are the key enzymes in the extrinsic and intrinsic coagulation cascades (Furie and Furie, 1988). When interacted with tissue factor (TF), FVII is converted to the active state (FVIIa), forms TF–FVIIa complex, and initiates the downstream extrinsic clotting cascade (Bogdanov et al., 2003). In the case of the intrinsic pathway of coagulation, FXII initiates the activation of the blood coagulation zymogen FXI and leads to the formation of factor IXa (Schmaier, 2008). Thus, further exploration of FVII and FXII may provide an in-depth insight into the aberrant coagulation in sepsis.
The circadian clock genes in mammals are endogenous oscillators, containing interlocked and negative feedback loops (Takahashi et al., 2008; Duong et al., 2011). These loops are basic helix-loop-helix-PER-ARNT-SIM (PAS) transcriptional activators BMAL1 and CLOCK, NPAS2 in addition to Period (Per1 and Per2), and cryptochrome (Cry1 and Cry2) genes (Koike et al., 2012). Among these genes, BMAL1 is a non-redundant and an essential component of circadian clock genes for maintaining normal physiology and behaviors (Bunger et al., 2000). Additionally, deficiency of BMAL1 in mice causes an imbalance in the coagulation mechanism and has potential for blood thrombin generation (Ninivaggi et al., 2014; Hemmeryckx et al., 2019), while other circadian clock genes such as CLOCK (Ohkura et al., 2006; Cheng et al., 2011), Per2 (Oishi et al., 2009), and Cry1/2 (Ohkura et al., 2006) participate in hemostasis via regulation of fibrinolytic systems rather than coagulation systems. Nevertheless, the mechanisms underlying the functional role of BMAL1 in affecting coagulation in bacterial infection, such as S. oralis, remain largely obscure.
In this study, we demonstrated that S. oralis downregulates the expression of BMAL1 and then disrupts the biosynthesis of coagulation factors. As a transcription factor, BMAL1 regulates the coagulation genes through the E-box elements. For the first time, this study demonstrated the correlation between S. oralis and BMAL1. The formation of microthrombus in hematosepsis is related to the balance of abnormal coagulation factors from hepatocyte due to the decline in BMAL1. Therefore, recovering BMAL1 expression might be a promising strategy to inhibit abnormal coagulation for reducing the formation of microthrombus and lowering the mortality.
Materials and Methods
Cell Culture and S. oralis Infection
HepG2 cells were purchased from ATCC (HB-8065, USA) and cultured in Dulbecco's minimum essential media (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 μg/mL streptomycin in an atmosphere of 5% CO2 at 37 °C. At 70–80% confluency, the cells were trypsinized and challenged by S. oralis with a multiplicity of infection (MOI)=50 for 24 h.
Animals
C57BL/6J mice were bought from the Beijing HFK Bioscience Co. Ltd (Beijing, China) and grouped randomly into either LD12:12 or jet-lagged. Homozygous BMAL1-deficiency (Bmal1−/−) mice in the C57BL/6J background were produced by breeding heterozygous BMAL1-deficiency mating pairs (Bmal1+/−), which was a kind gift from Dr. Ying Xu (Soochow University, Jiangsu, China). T BMAL1-deficiency was confirmed by Western blot as described (Bunger et al., 2000).
RNA Isolation and Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR)
Total RNA was extracted on ice using TRIzol (Vazyme, China), according to the manufacturer's protocols. cDNA synthesis was performed with reverse transcription reagent (Vazyme). Real-time RT-PCR was performed using SYBR Green PCR protocol on an ABI 7300 real-time PCR system (Applied Biosystems, Carlsbad, CA, USA). Relative mRNA expression of the target genes was normalized against that of GAPDH using the 2−ΔΔCt method, and presented as mean±SD of replicates. The primers used for amplification are listed in Supplementary Table 1.
Western Blot Analysis
Cells were lysed on ice by modified RIPA buffer (P0013B; Beyotime, Shanghai, China) to obtain total protein extract that was quantitated by BCA protein Assay kit (P0010; Beyotime, Shanghai, China), and resolved by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Subsequently, the proteins were transferred to polyvinylidene fluoride (PVDF) membranes that were then blocked in 5% milk diluted with TBST for 1 h at room temperature, probed with primary antibodies, such as BMAL1 (Abcam, 1:1000), CLOCK (Abcam, 1:1000), PER2 (Abcam, 1:1000), CRY2 (Abcam, 1:1000), GAPDH (Santa Cruz Biotechnology Inc., 1:10000), FVII (Santa Cruz Biotechnology Inc., 1:1000), and FXII (Santa Cruz Biotechnology Inc., 1:500), at 4°C overnight. Then, the membranes were incubated with a secondary goat anti-rabbit antibody (1:2000) on the following day for 1 h at room temperature. ECL enhanced chemiluminescence substrate kit (Millipore) was used for imaging and quantitation of the immunoreactive bands by the Image J software.
Immunohistochemistry (IHC)
Liver tissues were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) overnight and sliced into 5-μm-thick paraffin-embedded sections. After deparaffinization and rehydration, antigen retrieval was performed by boiling the sections in citrate antigen retrieval solution (P0081; Beyotime) for 15 min. Next, the endogenous peroxidase activity of the samples was blocked by 3% H2O2 for 15 min, followed by pre-blocking with 5% BSA (A1933; Sigma–Aldrich) for 1 h at room temperature. The levels of proteins were investigated in liver tissues using primary antibodies against BMAL1 (Abcam, 1:800), FVII (Santa Cruz Biotechnology Inc., 1:400), and FXII (Santa Cruz Biotechnology Inc., 1:400) using a Vectastain ABC kit (Vector Laboratories, Burlingame, CA, USA), followed by the DAB Substrate kit (Vector Laboratories). All protocols were followed according to the manufacturer's instructions.
Immunofluorescence (IF)
Cells were plated at a density of 1 × 105 cells/well in 24-well plates and then grown on glass coverslips for 12 h and infected with S. oralis, followed by staining with 5 mM SYTO® Green-Fluorescent Nucleic Acid Stains (Thermo Fisher, USA) for 30 min. After gentle washing for three times with PBS, the cells were fixed for 10 min in 4% paraformaldehyde (wt/vol). The cells were pre-blocked with 5% BSA (A1933; Sigma–Aldrich), labeled with TRITC Phalloidin (Yeasen, 1 μg/mL), and incubated with anti-rabbit Alexa Fluor 594 (Invitrogen) secondary antibodies (1:300) for 1 h at room temperature. The cell nuclei were stained with by DAPI dihydrochloride staining solution. Images were collected using a confocal microscope (SP2-AOBS Leica Microsystems).
Enzyme-Linked Immunosorbent Assay (ELISA)
The concentrations of mouse plasma FVII (Cusabio, China) and mouse plasma FXII (Cusabio) were measured using ELISA kits according to the manufacturer's instructions. HepG2 cells were seeded overnight in 1% FBS-DMEM in six-well plates (6 × 106/well) and infected with S. oralis in 100 U/mL penicillin and 100 μg/mL streptomycin for 24 h (MOI=50). Then, cell-free-culture supernatants were collected and analyzed for FVII or FXII content by a specific ELISA (), according to the manufacturer's recommendations. The cells without infection are similar to infected cells. Briefly, the standard protein was solubilized and diluted to 20, 10, 5, 2.5, 1.25, 0.625, and 0.312 ng/mL for constructing a standard curve. The diluted samples were added to the reaction plate and incubated at 37°C for 2 h. The wells were washed five times, and the biotin primary antibody was added, and the reaction was incubated at 37°C for 1 h. Subsequently, horseradish peroxidase (HRP)-avidin was added and incubated at 37°C for 1 h, followed by the addition of a substrate chromogenic solution that was incubated at 37°C for 30 min. Finally, the reaction terminator and OD450 were measured.
Fluorescence in situ Hybridization (FISH)
The following probe was used for FISH: S. oralis (probe sequences targeted 16S ribosomal-RNA (rRNA) genes; 6FAM-CTCCTACGGGAGGCAGCAGTAGGGA-BHQ-3; fluorescence emission maximum at 900 nM). As mentioned above, the liver sample was fixed in 4% paraformaldehyde for 2 h at room temperature and stored in 96% ethanol. Then, the paraffin-embedded sections (5-μm-thick) were incubated in hybridization buffer (100 mM Tris-HCl pH 7.2, 0.9 M NaCl, 0.1% SDS) containing 8 ng/mL of the S. oralis probe overnight at 37°C. Slides were washed 3 times for 15 min in prewarmed (37°C) hybridization buffer, following the labeling of the cell nucleus by DAPI. Images were collected using a confocal microscope (SP2-AOBS Leica Microsystems).
In vivo Imaging of Animals
For in vivo imaging, mice (LD12:12 and jet-lagged) were injected 109 fluorescence-labeled bacteria through the tail vein for 6 h prior to imaging, as described previously. Subsequently, the animals were euthanized with sodium pentobarbital, the hairs were wiped off, and the organs were removed and placed on plates. The fluorescence was detected at 490 nm (in-vivo FX PRO BRUKER). The images were analyzed using Bruker software. All procedures were performed using a standard protocol.
Coagulation Array
A volume of 1.8 mL heart blood was collected in a blood collection tube containing 0.2 mL of 3.2% sodium citrate anticoagulant (blood volume <1.8 mL was prepared according to the ratio of blood to anticoagulant 1:9) and mixed gently. The plasma was separated at 3,000 rpm for 15 min. The prothrombin time (PT), activated partial thromboplastin time (APTT), fibrinogen time, and fibrinogen concentration assays were conducted using a specific kit (Hanlisisw, China). All assays were carried out with an automatic coagulator (BE, Germany).
Bacterial Strains and Culture Conditions
The strain used for research: S. oralis from ATCC. Brain Heart Infusion (BHI) medium (Becton, Dickinson and Company, USA) was used for bacterial growth under aerobic conditions for 24–48 h at 37°C in order to reach the exponential growth phase. Subsequently, 10-fold serial dilutions of each bacterium were performed in PBS to achieve concentrations of ~1 × 108 CFU/mL. Specific primers were designed for S. oralis (forward primer: CAACGATACATAGCCGACCTGAG; reverse primer: TCCATTGCCGAAGATTCC).
Chromatin Immunoprecipitation Assay (ChIP)
Chromatin immunoprecipitation assays were conducted using chromatin immunoprecipitation kit (Millipore), according to the manufacturer's protocol. Briefly, HEPG2 cells were fixed in 1% formaldehyde, cross-linked with glycine, and lysed. Cell lysates were sonicated into fragments and precleared with Protein A/G Agarose beads for 1 h. After centrifugation (1,000 rpm × 1 min at 4°C), 10% of the supernatants were stored as input, and the remaining supernatants were divided into two parts, one for detection by BMAL1 antibody and the remaining for IgG reactivity (negative control). The DNA was purified in a final volume of 50 mL and subjected to PCR. Specific primers were designed for FVII promoter (forward primer: GACCGTCTACCCCAGTGTTT; reverse primer: CGGCAGTCCACGTCATTTC).
Luciferase Reporter Assay
Bmal1-overexpression and Bmal1-knockout HepG2 cells were constructed and plated at a density of 5 × 105 cells/well in 12-well plates. These cells were transfected with the wild-type FVII promoter, its deletion mutant, its mutation with mutated BMAL1-binding site, and empty vector (PGL3). Furthermore, firefly luciferase reporter vectors (100 ng) and Renilla luciferase reporter vectors pRL-SV40 (10 ng; Promega, USA) were transfected into cells using Lipofectamine™ 3,000 transfection kit (Thermo Fisher, USA) for 48 h. Subsequently, firefly and Renilla luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega). The data were normalized by the firefly/Renilla ratio.
Transformation of S. oralis
Briefly, mid-logarithmic phase cultures of S. oralis were diluted 1:20 in complete medium (Brain Heart Infusion, Becton, Dickinson, and Co.) containing 1 mM calcium chloride, 0.2% BSA (Sigma Aldrich Ltd, Dorset, UK) and 100 ng/mL competence-stimulating peptide 1 (CSP1; Mimotopes, Clayton, Victoria, Australia). The transformation reactions were incubated for 90 s at 42 °C and placed on ice for 2 min. Then, the sensory bacteria were transferred to 6-well plate with ampicillin for 16 h. The primers were designed for GFP (forward primer: GACCGTCTACCCCAGTGTTT; reverse primer: CGGCAGTCCACGTCATTTC).
Statistical Analysis
All the statistical analyses were performed with GraphPad Prism Software and based on normally distributed datasets with equal variance (Bartlett's test). The investigators were not blinded during the experiments or outcome assessment. The data points were presented as mean ± SD values unless otherwise stated. Data were inferred as statistically significant if P-values were <0.05. Significance between two groups was determined by independent Student's t-test. For multiple comparisons, one-way analysis of variance (ANOVA) was used with Tukey's multiple comparisons post-hoc test unless stated otherwise.
Result
S. oralis Reduced BMAL1 Expression and Disrupted the Biosynthesis of Coagulation Factors in vitro
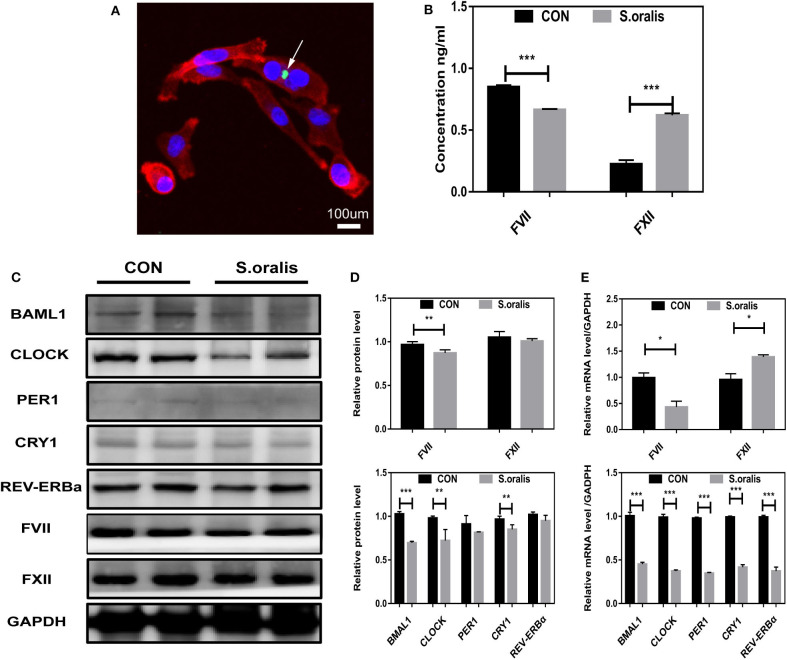
α-hemolytic Streptococcus, such as S. oralis, exhibits strong pathopoiesia either by generating various enzymes and exotoxin indirectly or invading into a host directly. In order to elucidate how S. oralis interacts with hepatocytes, nucleic acid dye-treated bacteria had infected cells in MOI=10,000 for 30 min, which showed that S. oralis directly invaded into the cells (Figure 1A). Owning to the lack of elements initiating coagulation, the clotting factors were not consumed, and hence, it was found that FVII slightly decreased, and FXII increased significantly in the cellular supernatant of infected HepG2 (Figure 1B). To explore the correlation between S. oralis infection and circadian clock genes in hepatocytes, cells were treated with S. oralis, and the total protein and mRNA were extracted. Consequently, BMAL1 expression was significantly decreased as compared to the other circadian clock genes, and it was found that the coagulation factor FVII decreased slightly, while the FXII did not alter (Figures 1C,D). In another cohort of cells with the same treatment, the expression of BMAL1 and other circadian clock genes was lower in S. oralis infection group, and obvious upregulation of FVII and mild downregulation of FXII was detected at the transcriptional level (Figure 1E).
Figure 1.
S. oralis reduced BMAL1 expression and disrupted the biosynthesis of coagulation factors in vitro. (A) Confocal images for HepG2 cells infected with S. oralis for 30 min, stained for DAPI (blue), F-actin (red), and S. oralis (green). The white arrow represents bacteria invading into cells. Scale bars, 100 μm. (B) Relative supernatant concentration of coagulation factors VII and XII in HepG2 cells infected with S. oralis for 48 h as compared to the uninfected group (CON). (C,D) Western blot analysis of circadian clock genes and coagulation factors, and the relative values of densitometry analysis. (E) qRT-PCR analysis of circadian clock genes and coagulation factors. The experiments were performed three times independently. *P < 0.05; **P < 0.01; ***P < 0.005.
S. oralis Reduced BMAL1 Expression and Disrupted the Biosynthesis of Coagulation Factors in vivo
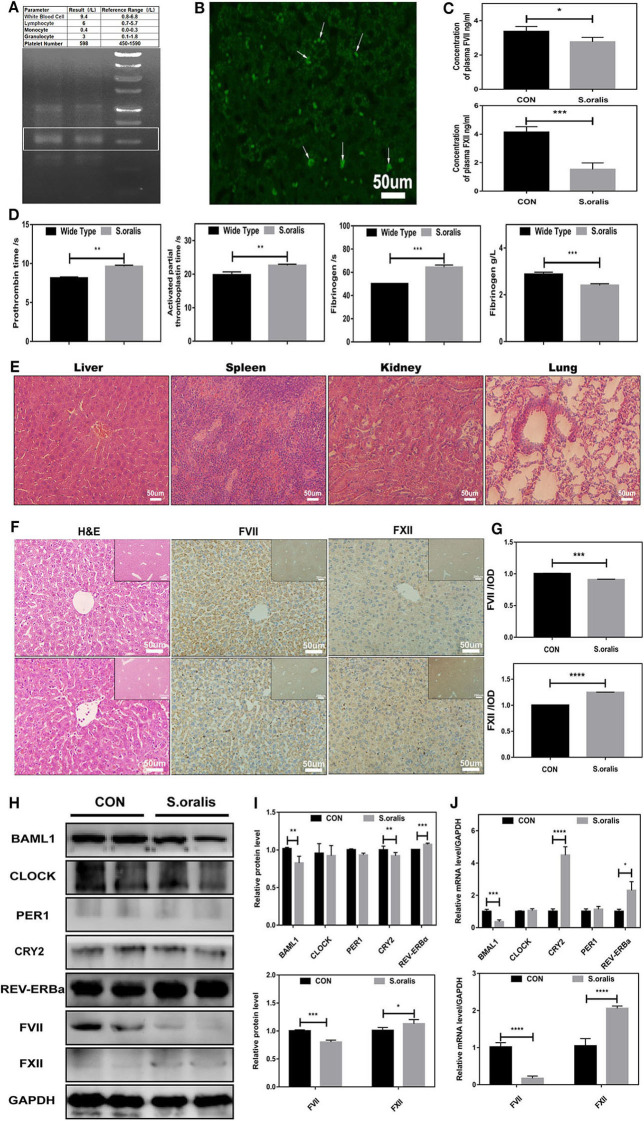
Correspondingly, to explore whether S. oralis modified the circadian clock genes and coagulation factor levels in vivo, another series of studies were carried out. The mice were injected with S. oralis into the tail vein. After the mice were sacrificed, the whole blood was collected for a routine examination, and it was found that the neutrophils increased significantly. In addition, the total DNA of the liver and spleen was extracted and amplified to detect the bacteria in these organs using S. oralis-specific primers (Figure 2A). To further consolidate the presence of bacteria in the liver, FISH was conducted, which showed green EGFP-labeled-bacterium in the liver, and many of them had invaded into the hepatocytes (Figure 2B). This phenomenon might suggest that S. oralis plays a functional role both directly and indirectly. The remaining of the whole blood was collected for coagulation arrays and ELISA. It was observed that the level of both plasma coagulation factors, FVII and FXII, was downregulated (Figure 2C). Additionally, it was found that PT, APTT, and fibrinogen reaction time were prolonged and plasma fibrinogen was declined (Figure 2D). In sepsis, bacteria can activate both intrinsic and extrinsic coagulation by the cytokine response. The serial sections of liver, spleen, kidney and lung stained with hematoxylin and eosin (H&E) indicated inflammation after infection (Figure 2E). Moreover, most of the coagulation factors, including FVII and FXII were generated in the liver. To investigate the correlation between S. oralis infection and coagulation factors, serial sections of the liver tissues were stained with H&E, and histochemistry was conducted (Figure 2F). S. oralis infection declined the expression of FVII expression, while that of FXII was elevated (Figure 2G). In addition, the livers of mice were collected and the total proteins were extracted. The results showed that BMAL1 expression was significantly decreased as compared to that of the other genes; also, an obvious decline was detected in FVII and a mild elevation of FXII as compared to the uninfected group (Figures 2H,I). On the other hand, BAML1 level was also decreased, while the mRNA expression of REV-ERBa and CRY1 was elevated, FVII was significantly downregulated, and FXII was markedly upregulated (Figure 2J). Therefore, we confirmed that S. oralis infection declined the level of FVII and BMAL1 while that of FXII was elevated at both protein and transcriptional levels in vivo.
Figure 2.
S. oralis reduced BMAL1 expression and disrupted the biosynthesis of coagulation factors in vivo. Every mouse was infected by S. oralis in about 1 × 108 CFU (colony-forming units) for 24 h. (A) The upper part for the blood routine test of the infected mouse, the lower part for DNA extraction of infected mouse livers. Special primers were designed for S. oralis. (B) FISH for the detection of S. oralis in infected mouse livers. Special primers were designed for S. oralis. Scale bars, 50 mm. (C) Plasma concentration of coagulation factors VII and XII in the infected group as compared to the control group. (D) Routine coagulation array for mice in infected and uninfected groups. (E) H&E staining for liver, spleen, kidney, and lung of infected mice. (F,G) IHC staining for coagulation factors VII and XII of the liver and the relative values of IHC optical density. (H,I) Western blot analysis of circadian clock genes and coagulation factors, and the relative values of densitometry analysis of hepatocytes. (J) qRT-PCR analysis of circadian clock genes and coagulation factors. The experiments were performed three times independently. *P < 0.05; **P < 0.01 ***P < 0.005; ****P < 0.001.
Downregulation of FVII and Upregulation of FXII in Bmal1-/- Mice
To confirm the role of BMAL1 in coagulation factor biosynthesis, we constructed BMAL1-deficiency (Bmal1−/−) mice, and the genotypes were verified by Western blot (Figure 2D). Interestingly, we found that BMAL1 had two-aspect functions in the coagulation process. The blood coagulation arrays showed that PT was slightly deduced while APTT was significantly prolonged with a huge reduction in these fibrinogen reaction time owning to elevated plasma fibrinogen in Bmal1−/− mice (Figure 3A). FVII and FXII were the key enzymes that initiated the extrinsic and intrinsic coagulation cascade, respectively. Therefore, the serial sections of the liver tissues were stained with H&E, and histochemistry was performed (Figure 3B). The result showed that FVII approximately decreased to a third, and FXII increased nearly four-fold in Bmal1−/− mice (Figure 3C). To further verify the correlation between BMAL1 and the biosynthesis of coagulation factors, the total protein of the liver tissues was extracted. This phenomenon was similar to that FVII decreased significantly while FXII increased by three-fold in Bmal1−/− mice (Figures 3D,E). In addition, that the expression of the coagulation factor was altered at the mRNA level, which was synchronous with the changes in the protein level, indicating that BMAL1 was likely to act as a transcription factor to regulate the biosynthesis of coagulation factors (Figure 3F). Although coagulation factors FVII and FXII played major roles in coagulation cascades, other coagulation factors, such as vWF, also played a significant role. Therefore, we analyzed the expression of other coagulation factors (Figure 3G). To investigate the effect of BAML1 on the biosynthesis of coagulation factors in vitro, Bmal1-overexpression or BAML1-knockout was used (Figure 3H), which showed that FVII expression increased mildly and FXII significantly decreased in Bmal1-overexpressing cells. However, a contrast result was observed in BAML1-knockout cells (Figure 3I). It was also confirmed that the deficiency of BMAL1 negatively regulated FVII expression but positively regulated FXII.
Figure 3.
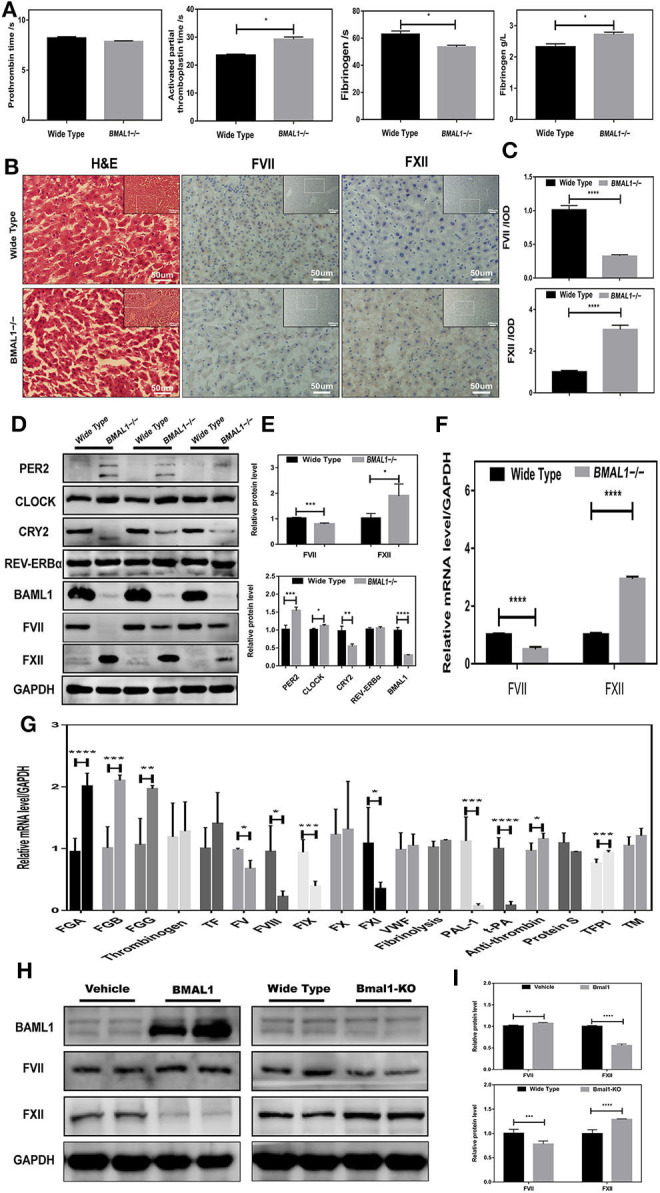
Disrupted coagulation factors VII and XII in Bmal1−/− mice. (A) Routine coagulation array for whole blood in Bmal1−/− vs. WT mice (3/group). (B,C) H&E and IHC staining for coagulation factors VII and XII in slices of livers, and the relative values of IHC optical density. (D–G) Western blot analysis and qRT-PCR analysis of circadian clock genes and coagulation factors in mouse liver. (H,I) Western blot analysis of coagulation factors VII and XII in Bmal1-overexpression and Bmal1-knockout HepG2 cells. The experiments were performed three times independently. *P < 0.05; **P < 0.01; ***P < 0.005; ****P < 0.001.
BMAL1 Regulates Coagulation Factors VII in a Direct Way
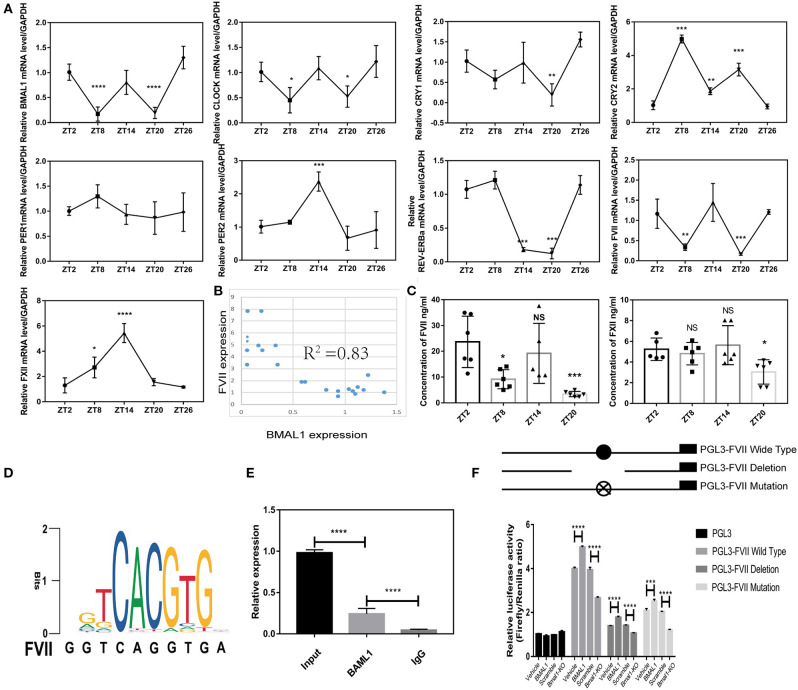
Firstly, we found that FVII mRNA level showed 12-h circadian changes and reached nadir around ZT8 and ZT20, but FXII did not exhibit circadian changes; several circadian clock genes in the liver showed obvious rhythmicity, and BMAL1 mRNA expression was synchronous with FVII expression by hitting valleys around ZT8 and ZT20 (Figure 4A). Furthermore, the correlation between circadian clock genes and FVII showed that the expression of BMAL1 was strongly associated with that of FVII (Figure 4B). To further verify the rhythmicity of FVII expression, we measured the plasma coagulation factor at different time points. Strikingly, FVII expression reached nadir around ZT8 and ZT20, while that of FXII lacked circadian changes (Figure 4C). Typically, BMAL1 acts as a transcription factor that forms a heterodimer with CLOCK to activate the expression of the downstream genes, such as E-box-controlled genes PER1 and CRY1 (Duong et al., 2011). Therefore, it could be deduced that BMAL1 regulated FVII through transcription factor binding sites (Figures 4D,E).
Figure 4.
BMAL1 regulates coagulation factors VII in a direct way. (A) Relative expression of circadian clock genes and coagulation factors at different time points (4 mice/each point). (B) Correlation analysis between BMAL1 and FVII. (C) Plasma concentration of coagulation factors VII and XII at each time point. (D) Motif sequence signature of BMAL1 in the promoter region of coagulation factor FVII. (E) ChIP assays were performed using an antibody against BMAL1, and IgG as negative controls. Specific primers were designed for FVII promoter. (F) Luciferase reporter assays were performed to measure the activities of WT FVII promoter (PGL3- FVII WT), its deletion mutant (PGL3- FVII deletion), its mutant (PGL3- FVII mutation) with mutated BMAL1-binding site and empty vector (PGL3) in Bmal1-overexpression and Bmal1-knockout HepG2 cells. The experiments were performed three times independently. *P < 0.05; **P < 0.01; ***P < 0.005; ****P < 0.001.
Low activity of FVII promoter could be found in cells transfected with both the FVII promoter mutants with Bmal1 binding site and the FVII promoter deletion mutant. To confirm the role of BMAL1, firefly luciferase reporter vectors were transfected into Bmal1-overexpressing and Bmal1-knockout cells. Next, we found that the activity of FVII promoter was enhanced in Bmal1-overexpressing cells, while it was declined in Bmal1-knockout cells. Together, these results suggested that BMAL1 directly regulates the expression of FVII in a positive way (Figure 4F).
Circadian Rhythm Disruption-Driven Coagulation Prevents Extravasation of S. oralis
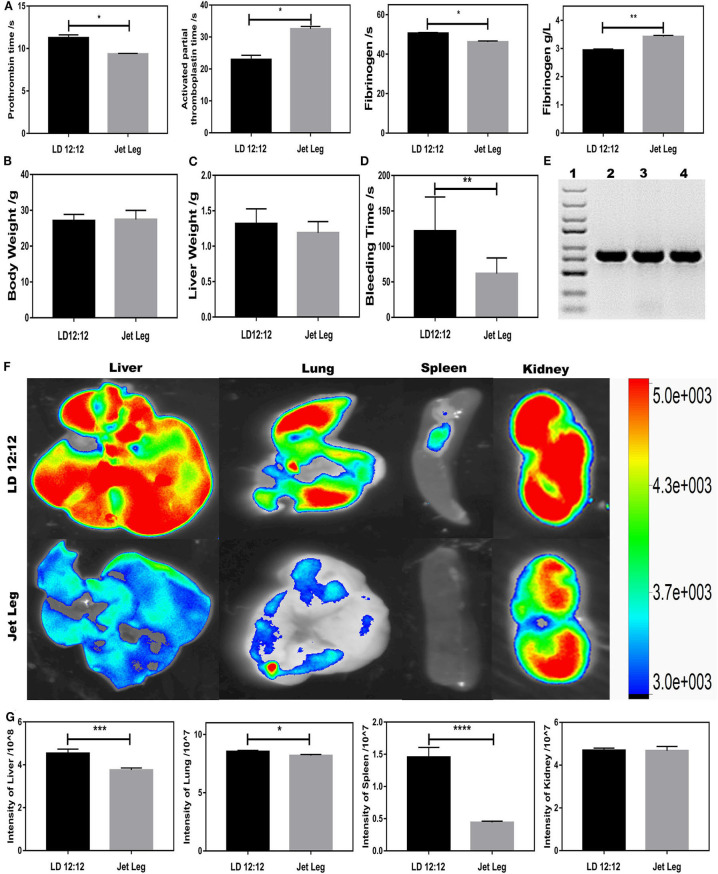
Since BMAL1 plays a key role in circadian rhythm, investigating the circadian rhythm disruption-driven coagulation is crucial for unveiling the role of BMAL1 in coagulation. To further explore the roles of circadian clock gene BMAL1 in coagulation, a jet-lagged mouse model was established to observe the impact of circadian rhythm disruption on the impairment of the liver ossification process. All mice were maintained under 12/12-h light/dark cycles with the light on from 8:00 a.m. (Zeitgeber time 0, ZT0) to 8:00 p.m. (ZT12) and fed antibiotic-free food and water ad libitum. In the experiment, C57BL/6J mice were placed on a previously described jet-lagged schedule with 8-h light advanced every 2–3 days for 12 weeks, which mimicked the circadian disturbance that humans undergo during shift work (Zhao et al., 2017). PT was mildly shortened, while APTT was significantly prolonged, and the fibrinogen reaction time was shortened owing to elevated plasma fibrinogen in jet-lagged mice (Figure 5A). To eliminate the impact of body weight and liver weight, measurement experiments were essential, which seemed to have no impact on the coagulation (Figures 5B,C). However, jet-lag seemed to induce a slight decline in BT, suggesting that circadian rhythm disruption-driven downregulation of BMAL1 might be conducive to thrombus formation (Figure 5D). In order to elucidate whether circadian rhythm disruption-driven coagulation prevents extravasation of S. oralis, EGFP-labeled S. oralis was constructed and confirmed by PCR amplification (Figure 5E). We found that the number of bacteria invaded into the liver was less than that in the LD12:12 group due to disruption-driven coagulation, which prevents the invasion of EGFP-labeled S. oralis. Intriguingly, a similar phenomenon was observed in the lung and spleen, and that the number of bacteria invaded into the kidneys was large in quantity in both the jet-lagged and LD12:12 groups because of abundant blood vessels (Figures 5F,G).
Figure 5.
Circadian rhythm disruption-driven coagulation prevents extravasation of S. oralis. (A) Routine coagulation array (4 mice/group) for whole blood in LD12:12 vs. jet-lag mice at ZT12. (B,C) Bodyweight and liver weight of mice in LD12:12 vs. jet-lag at ZT12. (D) Tail bleeding time of mice. (E) DNA extraction of EGFP-labeled S. oralis. Specific primers for GFP. (F,G) Imaging of mice in LD12:12 vs. jet-lag mice at ZT2 in vivo (4 mice/group). *P < 0.05; **P < 0.01; ***P < 0.005; ****P < 0.001.
Discussion
To gain new insight into the mechanism underlying the influence of the circadian gene BMAL1 on coagulation factors in bacterial infectious liver disease, we performed the experiment described above and showed that the level of BMAL1 in hepatocytes was downregulated in the S. oralis infection group, leading to the downregulation of coagulation factor VII and the upregulation of coagulation factor XII. Similar results were found in vitro. Bmal1 overexpression significantly increased the coagulation factor VII while it decreased the level of coagulation factor XII in HEPG2 cell line. According to these findings, circadian gene BMAL1 plays a critical role in coagulation by regulating the biogenesis of clotting factors in S. oralis infection.
Reportedly, the gut homeostatic microbiome is necessary for normal oscillations in both intestine and liver, which is effectuated via circadian genes, and participating in diurnal fluctuations of host physiology, disease susceptibility, lipid absorption, and sleep rhythms (Miyazaki et al., 2014; Thaiss et al., 2016; Kuang et al., 2019). The courses of bacterial invasion into the host include bacterial adsorption on the body surface, invading tissues or cells, growing and breeding, producing toxins, spreading, and resisting a series of defensive functions of the host. Colonized S. oralis derived from blood bacteria was found in the liver and was also found to be invading into hepatocytes (Figures 1A, 2A). Herein, we observed that the downregulation of BMAL1 contributed to the secretion of FXII without the upregulation of FXII protein level in vitro (Figure 1B). Consistent with the current findings, some studies have shown that the deficiency of BMAL1 upregulates the secretion of many biochemical parameters by restraining their repressors, such as CXCL5 (Gibbs et al., 2014) and interleukin-1β (Luyts et al., 2014). Furthermore, the interaction between Bmal1 of hepatocytes and bacterial infection revealed that the expression of BMAL1 descended at the transcriptional level both in vitro and in vivo in the microorganism. This finding is in line with the results of the previous studies (Lou et al., 2017; Li et al., 2018; Coiffard et al., 2019). With the infection of Mycobacterium tuberculosis, the level of BMAL1 was significantly decreased in both the lung and the spleen (Lou et al., 2017). Conversely, some studies showed a certain promoting effect of BMAL1 on gastric epithelial cells with the infection of Helicobacter pylori (Li et al., 2018), which could be attributed to the various roles of BMAL1 in different diseases or the various types of bacteria executing infections in different conditions. The reciprocal role between bacteria and BMAL1 has been reported. The magnitude of response to the pathogenic bacterial challenge under the control of Bmal1 has been established (Bhardwaj et al., 2011; Wang et al., 2011; Gibbs et al., 2012, 2014). However, the mutual role of Bmal1 and microbiota needs to be investigated in the future.
Circadian rhythm is a regular recurrence of biological processes or activities with an ~24-h cycle. The circadian changes have been detected in human coagulation factors, such as antithrombin, FVII, and FVIII (Kapiotis et al., 1997; Undar et al., 1999; Iversen et al., 2002). In the current study, not only mouse plasma FVII but also FVII in mouse hepatocyte exhibit rhythmicity (Figure 4A). BMAL1 plays a core and elemental role (Bunger et al., 2000). However, other components of circadian clock genes, such as CLOCK and CRY1/2 (Ohkura et al., 2006; Cheng et al., 2011), were largely relevant to the fibrinolytic system rather than the coagulation system. The properties of diurnal rhythm were entirely lost for FVII and antithrombin. Nonetheless, additional evidence on the regulation of coagulation factors needs to be unveiled in Bmal1−/− mice. This study showed that FVII level was tremendously declined in Bmal1−/− mice (Figures 3C,E). Interestingly, FXII level was markedly elevated in Bmal1−/− mice (Figures 3C,E). Thus, our findings may be a significant breakthrough that BMAL1 plays a two-sided role in regulating the coagulation process. In the endogenous coagulation pathway, decreased FXI and FXI might explain the prolonged APPT despite the elevated level of FXII in Bmal1−/− mice. In the exogenous coagulation pathway, deficiency of BAML1 might explain the unchanged PT by affecting the activation of coagulation factors despite deduced FVII in Bmal1−/− mice (Figure 3G). This displays that the regulation of FVII is complicated and sophisticated. BMAL1 is crucial for the regulation of FVII expression. Low expression of BMAL1 could still be found in Bmal1-knockout cells (Figure 3G). These phenomena might explain the low activity of FVII promoter, which could be found in Bmal1-knockout cells (Figure 4F). In the future, it might be essential to study the correlation between BMAL1 and coagulation.
The elevated FXII level ascribes the potentially elevated generation of endogenous thrombin larger in BMAL1-deficient mice (Hemmeryckx et al., 2019). Additionally, the level of FGA (Fibrinogen α chain), FGB (Fibrinogen β chain), and FGG (Fibrinogen γ chain) was elevated in Bmal1−/− mice in accordance with the elevated plasma fibrinogen in the blood coagulation arrays, which contributed to the formation of the crypto-fibrin network (Figure 3G; Ninivaggi et al., 2014). Although circadian rhythm disruption-driven coagulation prevents the extravasation of S. oralis, more macrothrombusus is induced than in the control group, which leads to multiple organ failure on account of deduced BMAL1 expression.
In summary, our results demonstrated that the downregulation of BMAL1 by S. oralis infection disrupts these clotting factors in hepatocytes. Therefore, we identified the coagulation factors VII and XII that were largely regulated in Bmal1−/− mice. Interestingly, this study on circadian clock genes BMAL1 might provide a novel view for the generation of microthrombus in hematosepsis induced by S. oralis.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by the Committee of Ethics of Wuhan Union Hospital and the Institutional Research Ethics Committee of Tongji Medical College, Huazhong University of Science and Technology (Wuhan, China). Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
LC and SL designed the study and critically revised the manuscript. JN performed the experiments, analyzed the data, and drafted the manuscript. JZ participated in the implementation of the study. SY, YL, and JP collected the data. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Ying Xu for the heterozygous BMAL1-deficient mating pairs (Bmal1wt/−).
Footnotes
Funding. This work was financially supported by grants from the National Science Foundation for Distinguished Young Scholars of China (31725011 to LC) and the National Natural Science Fund (81700946 to JZ).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2020.530190/full#supplementary-material
References
- Amaral A., Opal S. M., Vincent J.-L. (2004). Coagulation in sepsis. Intensive Care Med. 30, 1032–1040. 10.1007/s00134-004-2291-8 [DOI] [PubMed] [Google Scholar]
- Bhardwaj V., Meier S., Petersen L. N., Ingle R. A., Roden L. C. (2011). Defence responses of Arabidopsis thaliana to infection by Pseudomonas syringae are regulated by the circadian clock. PLoS ONE 6:e26968. 10.1371/journal.pone.0026968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanov V. Y., Balasubramanian V., Hathcock J., Vele O., Lieb M., Nemerson Y. (2003). Alternatively spliced human tissue factor: a circulating, soluble, thrombogenic protein. Nat. Med. 9, 458–462. 10.1038/nm841 [DOI] [PubMed] [Google Scholar]
- Bunger M. K., Wilsbacher L. D., Moran S. M., Clendenin C., Radcliffe L. A., Hogenesch J. B., et al. (2000). Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 103, 1009–1017. 10.1016/S0092-8674(00)00205-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S., Jiang Z., Zou Y., Chen C., Wang Y., Liu Y., et al. (2011). Downregulation of Clock in circulatory system leads to an enhancement of fibrinolysis in mice. Exp. Biol. Med. 236, 1078–1084. 10.1258/ebm.2011.010322 [DOI] [PubMed] [Google Scholar]
- Coiffard B., Diallo A. B., Culver A., Mezouar S., Hammad E., Vigne C., et al. (2019). Circadian rhythm disruption and sepsis in severe trauma patients: Shock 52, 29–36. 10.1097/SHK.0000000000001241 [DOI] [PubMed] [Google Scholar]
- Cornely O. A., Schirmacher P. (2001). Clinical picture: bacterial translocation in neutropenic sepsis. Lancet. 358:1842. 10.1016/S0140-6736(01)06884-2 [DOI] [PubMed] [Google Scholar]
- Duong H. A., Robles M. S., Knutti D., Weitz C. J. (2011). A molecular mechanism for circadian clock negative feedback. Science 332, 1436–1439. 10.1126/science.1196766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourrier F., Chopin C., Goudemand J., Hendrycx S., Caron C., Rime A., et al. (1992). Septic shock, multiple organ failure, and disseminated intravascular coagulation. Compared patterns of antithrombin III, protein C, and protein S deficiencies. Chest 101, 816–823. 10.1378/chest.101.3.816 [DOI] [PubMed] [Google Scholar]
- Furie B., Furie B. C. (1988). The molecular basis of blood coagulation. Cell 53, 505–518. 10.1016/0092-8674(88)90567-3 [DOI] [PubMed] [Google Scholar]
- Gibbs J., Ince L., Matthews L., Mei J., Bell T., Yang N., et al. (2014). An epithelial circadian clock controls pulmonary inflammation and glucocorticoid action. Nat. Med. 20, 919–926. 10.1038/nm.3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs J. E., Blaikley J., Beesley S., Matthews L., Simpson K. D., Boyce S. H., et al. (2012). The nuclear receptor REV-ERBα mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proc. Natl. Acad. Sci. U.S.A. 109, 582–587. 10.1073/pnas.1106750109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmeryckx B., Frederix L., Lijnen H. R. (2019). Deficiency of Bmal1 disrupts the diurnal rhythm of haemostasis. Exp. Gerontol. 118, 1–8. 10.1016/j.exger.2018.12.017 [DOI] [PubMed] [Google Scholar]
- Iversen P. O., Groot P. D., Hjeltnes N., Andersen T. O., Mowinckel M. C., Sandset P. M., et al. (2002). Impaired circadian variations of haemostatic and fibrinolytic parameters in tetraplegia. Br. J. Haematol. 119, 1011–1016. 10.1046/j.1365-2141.2002.03953.x [DOI] [PubMed] [Google Scholar]
- Kapiotis S., Jilma B., Quehenberger P., Ruzicka K., Handler S., Speiser W. (1997). Morning hypercoagulability and hypofibrinolysis. Diurnal variations in circulating activated factor VII, prothrombin fragment F1+2, and plasmin-plasmin inhibitor complex. Circulation 96, 19–21. 10.1161/01.CIR.96.1.19 [DOI] [PubMed] [Google Scholar]
- Koike N., Yoo S.-H., Huang H.-C., Kumar V., Lee C., Kim T.-K., et al. (2012). Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338, 349–354. 10.1126/science.1226339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang Z., Wang Y., Li Y., Ye C., Ruhn K. A., Behrendt C. L., et al. (2019). The intestinal microbiota programs diurnal rhythms in host metabolism through histone deacetylase 3. Science 65, 1428–1434. 10.1101/580613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Helmerhorst E. J., Leone C. W., Troxler R. F., Yaskell T., Haffajee A. D., et al. (2004). Identification of early microbial colonizers in human dental biofilm. J. Appl. Microbiol. 97, 1311–1318. 10.1111/j.1365-2672.2004.02420.x [DOI] [PubMed] [Google Scholar]
- Li T., Shao W., Li S., Ma L., Zheng L., Shang W., et al. (2018). H. pylori infection induced BMAL1 expression and rhythm disorder aggravate gastric inflammation. EBioMedicine 39, 301–314. 10.1016/j.ebiom.2018.11.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou J., Wang Y., Zhang Z., Qiu W. (2017). Activation of MMPs in macrophages by Mycobacterium tuberculosis via the miR-223-BMAL1 signaling pathway. J. Cell. Biochem. 118, 4804–4812. 10.1002/jcb.26150 [DOI] [PubMed] [Google Scholar]
- Luyts K., Smulders S., Napierska D., van kerckhoven S., Poels K., Scheers H., et al. (2014). Pulmonary and hemostatic toxicity of multi-walled carbon nanotubes and zinc oxide nanoparticles after pulmonary exposure in Bmal1 knockout mice. Part Fibre Toxicol. 11:16. 10.1186/s12989-014-0061-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki K., Itoh N., Yamamoto S., Higo-Yamamoto S., Nakakita Y., Kaneda H., et al. (2014). Dietary heat-killed Lactobacillus brevis SBC8803 promotes voluntary wheel-running and affects sleep rhythms in mice. Life Sci. 111, 47–52. 10.1016/j.lfs.2014.07.009 [DOI] [PubMed] [Google Scholar]
- Ninivaggi M., Kelchtermans H., Kuijpers M. J., Hemmeryckx B., Heemskerk J. W. M., Lindhout T., et al. (2014). Whole blood thrombin generation in Bmal1-deficient mice. Thromb. Haemost. 112, 271–275. 10.1160/TH13-11-0910 [DOI] [PubMed] [Google Scholar]
- Ohkura N., Oishi K., Fukushima N., Kasamatsu M., Atsumi G.-I., Ishida N., et al. (2006). Circadian clock molecules CLOCK and CRYs modulate fibrinolytic activity by regulating the PAI-1 gene expression. J. Thromb. Haemost. 4, 2478–2485. 10.1111/j.1538-7836.2006.02210.x [DOI] [PubMed] [Google Scholar]
- Oishi K., Miyazaki K., Uchida D., Ohkura N., Wakabayashi M., Doi R., et al. (2009). PERIOD2 is a circadian negative regulator of PAI-1 gene expression in mice. J. Mol. Cell. Cardiol. 46, 545–552. 10.1016/j.yjmcc.2009.01.001 [DOI] [PubMed] [Google Scholar]
- Penack O., Becker C., Buchheidt D., Christopeit M., Kiehl M., von Lilienfeld-Toal M., et al. (2014). Management of sepsis in neutropenic patients: 2014 updated guidelines from the infectious diseases working party of the German society of hematology and medical oncology (AGIHO). Ann. Hematol. 93, 1083–1095. 10.1007/s00277-014-2086-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmaier A. H. (2008). The elusive physiologic role of Factor XII. J. Clin. Invest. 118, 3006–3009. 10.1172/JCI36617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelburne S. A., Sahasrabhojane P., Saldana M., Yao H., Su X., Horstmann N., et al. (2014). Streptococcus mitis strains causing severe clinical disease in cancer patients. Emerg. Infect. Dis. 20, 762–771. 10.3201/eid2005.130953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi J. S., Hong H.-K., Ko C. H., McDearmon E. L. (2008). The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat. Rev. Genet. 9, 764–775. 10.1038/nrg2430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaiss C. A., Levy M., Korem T., Dohnalov,á L., Shapiro H., Jaitin D. A., et al. (2016). Microbiota diurnal rhythmicity programs host transcriptome oscillations. Cell 167, 1495–1510.e12. 10.1016/j.cell.2016.11.003 [DOI] [PubMed] [Google Scholar]
- Thiagarajan S., Krishnamurthy S., Raghavan R., Mahadevan S., Madhugiri V. S., Sistla S. (2016). Streptococcus oralis cerebral abscess following monkey bite in a 2-month-old infant. Paediatr. Int. Child Health 36, 160–162. 10.1179/2046905515Y.0000000020 [DOI] [PubMed] [Google Scholar]
- Turnier L., Nausheen S., Cunha B. A. (2009). Fatal Streptococcus viridans (S. oralis) aortic prosthetic valve endocarditis (PVE) with paravalvular abscesses related to steroids. Heart Lung 38, 167–171. 10.1016/j.hrtlng.2007.12.008 [DOI] [PubMed] [Google Scholar]
- Undar L., Ertugrul C., Altunba,ş H., Akça S. (1999). Circadian variations in natural coagulation inhibitors protein C, protein S and antithrombin in healthy men: a possible association with interleukin-6. Thromb. Haemost. 81, 571–575. 10.1055/s-0037-1614526 [DOI] [PubMed] [Google Scholar]
- Vervloet M. G., Thijs L. G., Hack C. E. (1998). Derangements of coagulation and fibrinolysis in critically III patients with sepsis and septic shock. Semin. Thromb. Hemost. 24, 33–44. 10.1055/s-2007-995821 [DOI] [PubMed] [Google Scholar]
- Wang W., Barnaby J. Y., Tada Y., Li H., Tör M., Caldelari D., et al. (2011). Timing of plant immune responses by a central circadian regulator. Nature 470, 110–114. 10.1038/nature09766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhou X., Tang Q., Yu R., Yu S., Long Y., et al. (2017). BMAL1 deficiency contributes to mandibular dysplasia by upregulating MMP3. Stem Cell Rep. 10, 180–195. 10.1016/j.stemcr.2017.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.