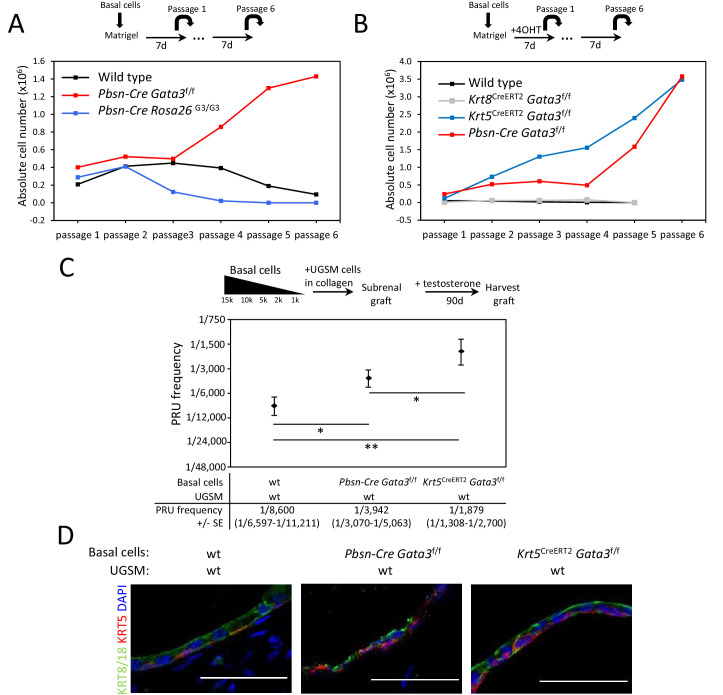
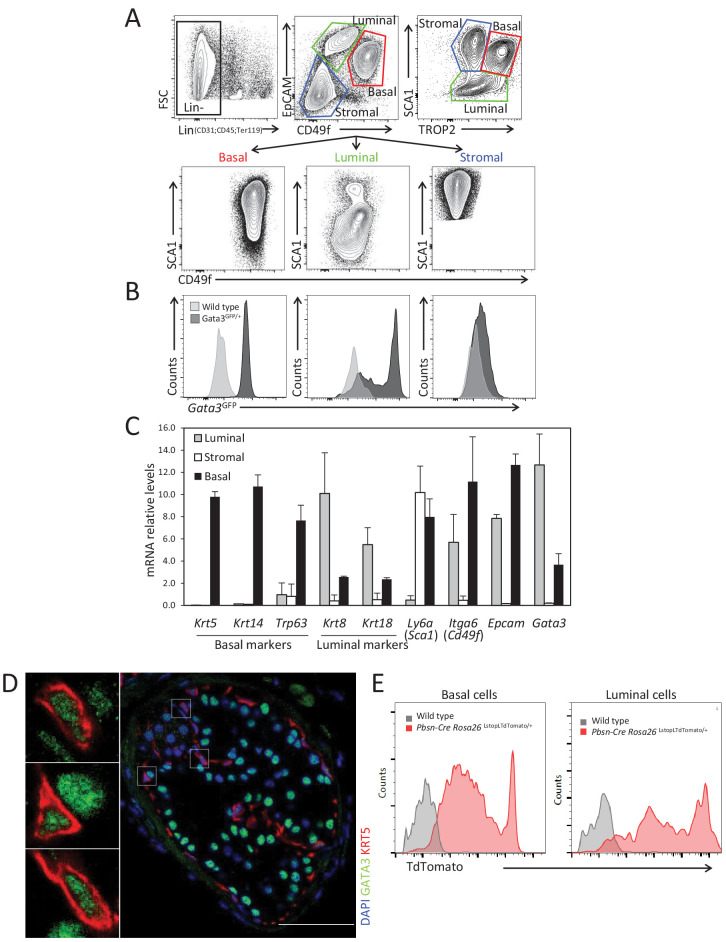
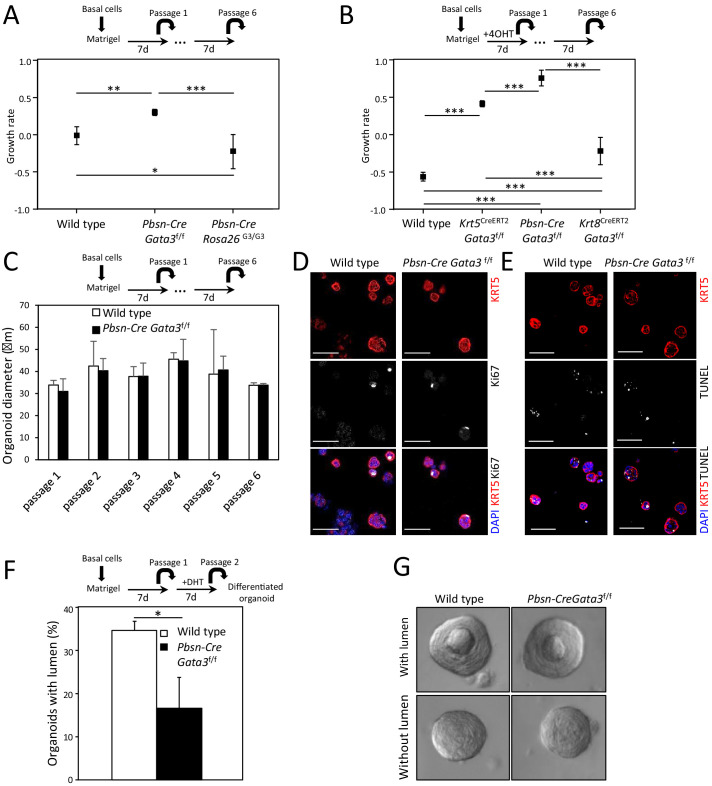
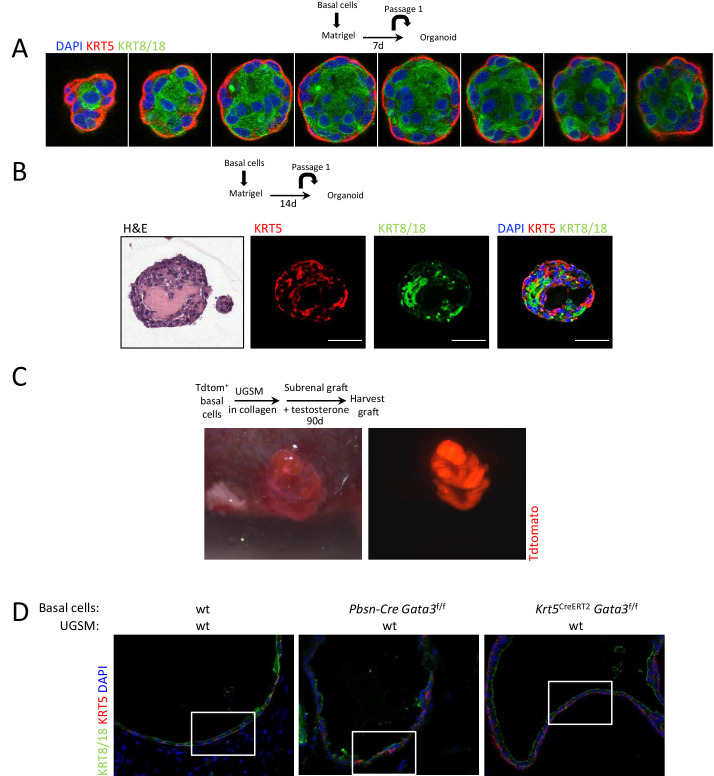
Figure 1. Gata3 loss leads to an expansion of prostate basal stem/progenitor cells numbers.
(A) Effect of Gata3 loss and overexpression on in vitro basal stem/progenitor maintenance potential. Organoid-forming potential was assessed by plating equal numbers (105 cells) of sorted basal prostate cells from the indicated genotypes in Matrigel and passaged every 7 days. Shown is the absolute number of cells obtained after each passage for the indicated genotypes. Data are representative of two independent experiments from a pool of prostate cells from a minimum of three mice. (B) Specific deletion of Gata3 in KRT5+ basal cells affect the organoid-forming potential upon passage. Organoid-forming potential was assessed as in (A). Cre activity was induced in vitro by treatment with hydroxy-tamoxifen for the first passage. (C) Gata3 loss increase regenerative capacity in vivo. Different numbers of sorted basal cells from wild type (Pbsn-Cre, Pbsn-Cre Gata3f/f and Krt5CreERT2Gata3f/f) prostate were mixed with UGSM and transplanted under the kidney capsule of immunodeficient mice. All mice contain Rosa26LstopLTdTomato/+ allele and Cre activity was induced in vivo by tamoxifen injection in adult mice 4 weeks prior to organoid propagation potential assessment. Prostate reconstituting units (PRU) frequency of total basal cells was calculated based on growth of TdTomato+ grafts using the Limiting Dilution Analysis software L-Calc (StemCell Technologies) according to Poisson statistics (two-tailed t-test; *p<0.05, **p<0.001). (D) Immunofluorescence staining of lineage-specific markers KRT5 (basal) and KRT8/18 (luminal) in wild type, Pbsn-Cre Gata3f/f and Krt5CreERT2Gata3f/f allografts. Scale bar is representative of 50 μm. See also Figure 1—figure supplements 1–2.