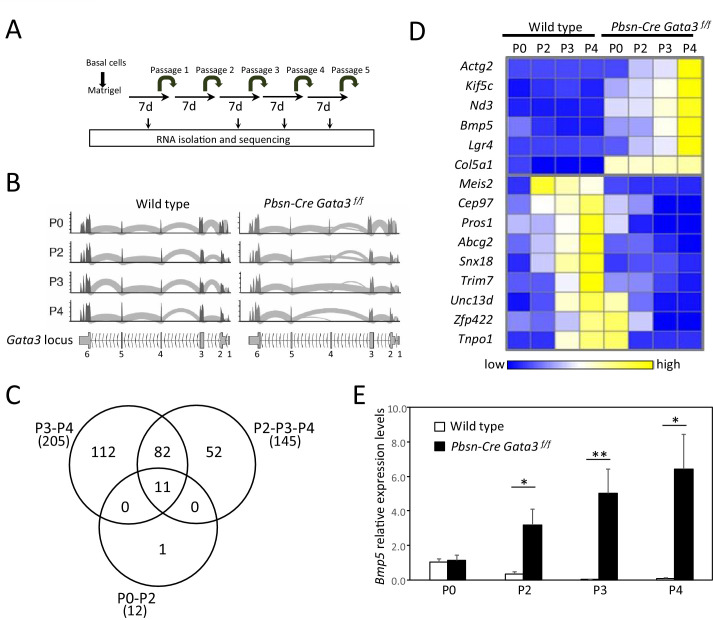
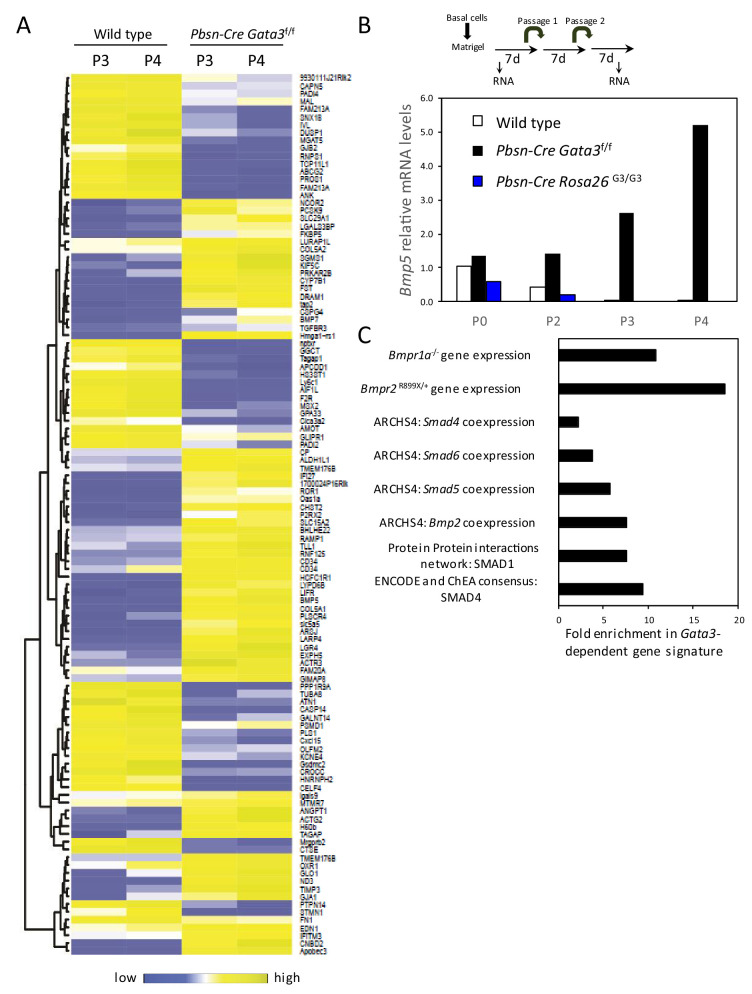
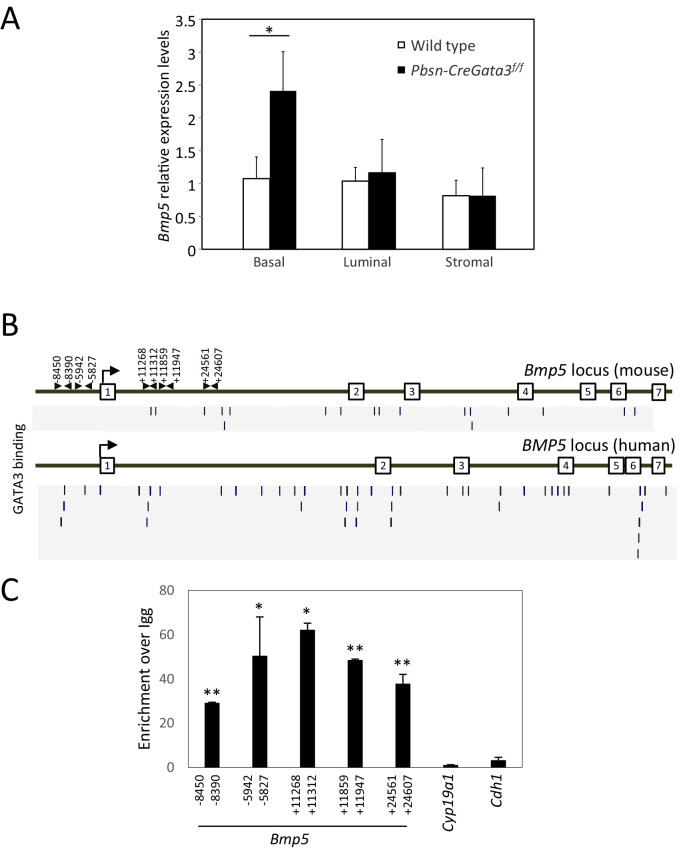
Figure 2. Bmp5 expression in organoids is regulated by Gata3.
(A) Schematic of RNA-seq strategy. mRNA was isolated from 4 days old wild type and Pbsn-Cre Gata3f/f organoids at passages P0, P2, P3 and P4. (B) Deletion of exon four in Pbsn-Cre Gata3f/f samples increases with passages. Shown are the read counts from RNAseq assigned to the Gata3 locus in samples isolated from wild type and Pbsn-Cre Gata3f/f prostate tissue at passage P0, P2, P3 or P4. (C) Venn diagram of genes differentially expressed between wild type and Pbsn-Cre Gata3f/f prostate organoids using likelihood-ratio test with q-value <0.01. (D) Heatmap of log2 transformed mRNA read counts of differentially expressed genes between wild type and Pbsn-Cre Gata3f/f organoids and whose expression pattern follows Gata3 loss with passages. (E) Bmp5 mRNA expression levels as assessed by quantitative RT-PCR in both wild type and Pbsn-Cre Gata3f/f organoids over passages. Data represent the average ± SD from three independent cDNA obtained from a pool of prostate cells from a minimum of three mice. Relative mRNA expression levels are normalized to Ppia mRNA levels (two-tailed t-test as compared to wild-type condition; *p<0.01, **p<0.005). See also Figure 2—figure supplements 1–2.