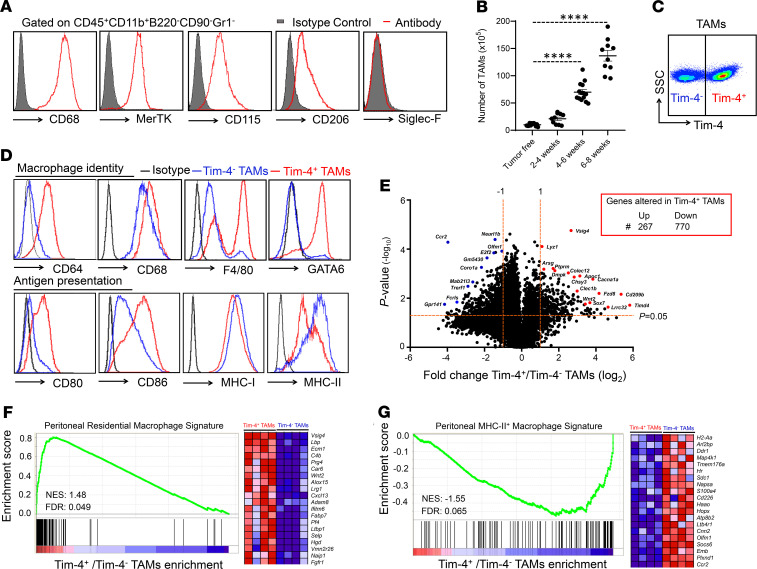
Figure 1. Tim-4 defines 2 distinct peritoneal macrophage subsets in ovarian cancer.
(A) Measurement of macrophage-related markers on TAMs. CD45+CD11b+B220–CD90–Gr1– macrophages were identified in peritoneal single cells in mice bearing peritoneal ID8 ovarian cancer (n = 10). (B) Dynamic changes of peritoneal TAMs during ID8 peritoneal ovarian cancer progression (n = 10–14/group, mean ± SEM). Time points: 2–4, 4–6, and 6–8 weeks. ****P < 0.0001 (1-way ANOVA with Dunnett’s multiple-comparisons test) between tumor-free and tumor-bearing mice. (C) Representative surface expression of Tim-4 on peritoneal TAMs. Data are shown at week 4 after ovarian cancer inoculation (n = 10). (D) Phenotypic difference in Tim-4+ and Tim-4- TAMs. TAMs were stained with the indicated antibodies. One representative of 5 is shown. (E) Transcripts in TAM subsets. Peritoneal TAM subsets were isolated and sorted from 6 to 7 weeks in ID8 tumor–bearing mice. An RNA-Seq assay was performed in 4 groups of paired Tim-4+ and Tim-4– TAMs. Volcano plots show upregulated and downregulated genes based on statistic value P < 0.05 and fold change ≥ 2 or ≤ –2. (F and G) RNA-Seq analysis in TAMs. Positive gene enrichment of residential macrophage gene signatures (F) and negative gene enrichment of MHC-II+ macrophage gene signatures (G) in Tim-4+ TAMs compared with Tim-4– TAMs. The 20 most enriched genes are shown on the right side. NES, normalized enrichment score; FDR, false discovery rate. FDR < 0.25 is considered significant.