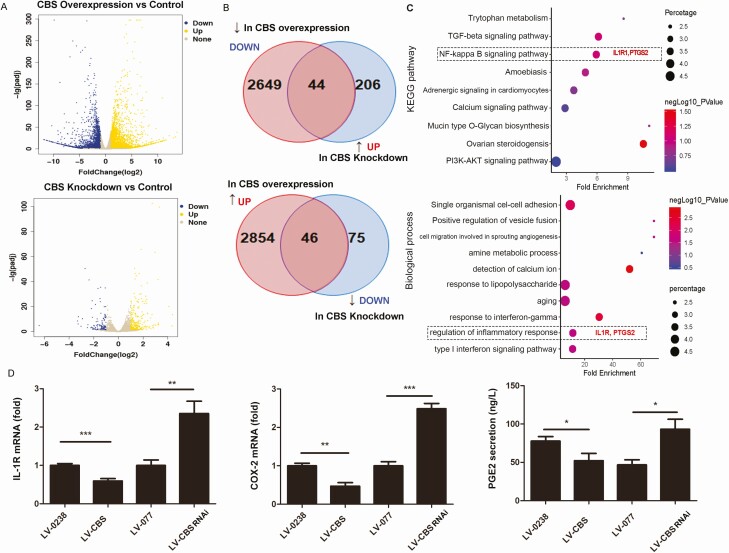
Figure 3.
Differentially expressed genes in HTR8/SVneo cells transfected with CBS siRNA or CBS cDNA.The transcriptomes of cells with CBS downregulation or upregulation were analyzed by RNA-seq analysis. Differentially expressed genes (DEGs) between CBS downregulation and upregulation versus control were presented in volcano plots (A) and Venn diagrams (B) and gene ontology (GO) analyses of top 9 KEGG pathways (C, upper panel) and top 10 biological processes (C, lower panel). GO analysis identified IL-1R and PTGS2 to be significantly associated with regulation of inflammatory response. IL-1R and PTGS2 (COX2) were further confirmed by qPCR (D), and PGE2 secretion was determined by ELISA (D). Data were calculated as means ± SD from at least 3 independent experiments. * P < 0.05; ** P < 0.01; *** P < 0.001. CBS, cystathionine β-synthase; cDNA, complementary deoxyribonucleic acid; COX2, cyclooxygenase-2; ELISA, enzyme-linked immunosorbent assay; IL-1R, interleukin-1 receptor 1; PGE2, prostaglandin E2; PTGS2, prostaglandin synthase 2; qPCR, quantitative polymerase chain reaction; RNA-seq, ribonucleic acid sequencing; siRNA, small interfering RNA.