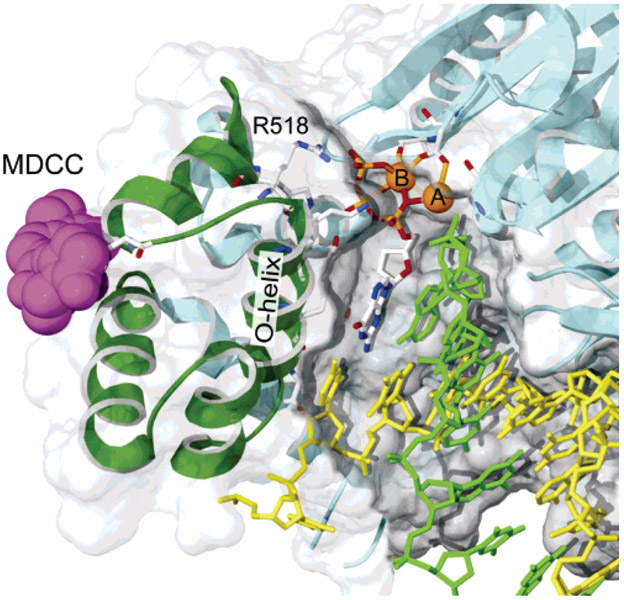
Figure 1:
CSF probe labeling. The structure of T7 DNA polymerase is shown with MDCC docked at the position expected for the labeling of C514. The thumb and thioredoxin-binding domain (residues 233–411) and primer binding loop (residues 436–454) have been removed to reveal the active site. From PDB 1T7P (14), a cys-light mutant was constructed by removing 8 of the 10 cysteines (C20S–C88A–C275A–C313A–C451S–C660A–C688A–C703A) and introducing a single-surface-exposed cysteine (E514C) and was labeled overnight at 4 °C with a 20-fold molar excess of MDCC. Only residue 514 was labeled, and 90% efficiency was achieved.