Reports of transmissible colistin resistance show the importance of comprehensive colistin resistance surveillance. Recently, a new allele of the mobile colistin resistance (mcr) gene family designated mcr-9, which shows variation in genetic context and colistin susceptibility, was reported. We tested over 100 Salmonella enterica and Escherichia coli isolates with mcr-9 from the National Antimicrobial Resistance Monitoring System (NARMS) in the United States for their susceptibility to colistin and found that every isolate was susceptible, with an MIC of ≤1 μg/ml.
KEYWORDS: colistin, genomics, PacBio, conjugation, mcr
ABSTRACT
Reports of transmissible colistin resistance show the importance of comprehensive colistin resistance surveillance. Recently, a new allele of the mobile colistin resistance (mcr) gene family designated mcr-9, which shows variation in genetic context and colistin susceptibility, was reported. We tested over 100 Salmonella enterica and Escherichia coli isolates with mcr-9 from the National Antimicrobial Resistance Monitoring System (NARMS) in the United States for their susceptibility to colistin and found that every isolate was susceptible, with an MIC of ≤1 μg/ml. Long-read sequencing of 12 isolates revealed mcr-9 on IncHI plasmids that were either independent or integrated into the chromosome. Overall, these results demonstrate that caution is necessary when determining the clinical relevance of new resistance genes.
INTRODUCTION
Colistin is an antimicrobial drug of last resort used to treat serious Gram-negative bacterial infections. It is not used as a first-line antimicrobial agent due to its toxicity, but it may be used to treat resistant infections (1). Colistin works by targeting the negatively charged membrane of Gram-negative bacteria, resulting in membrane disruption and cell death (2). Colistin has also been used as a growth promoter in food animals in Europe and Asia (3), until recent governmental actions restricted or banned these uses (4, 5). Colistin has not been used in food animals in the United States.
Colistin resistance has historically been rare, with known mechanisms being restricted to certain mutations, such as those in pmrA and pmrB (6). In 2015, the mobile colistin resistance gene mcr-1, conferring transmissible colistin resistance, was discovered (7). This gene has been found worldwide in many bacterial species, including Escherichia coli, Klebsiella pneumoniae, and Salmonella enterica, among others (8–10). In subsequent years, an additional eight mcr variants have been described and assigned new numerical designations (11, 12). Despite these reports, mcr gene prevalence in the United States has remained low, with most findings in human patients attributed to infections acquired as part of international travel (13, 14). Prior to this study, the National Antimicrobial Resistance Monitoring System (NARMS), a resistance surveillance program in the United States, had not identified any retail meat isolates with any mcr variant.
In 2019, the latest variant, mcr-9, was reported (15). This allele had only 64.5% amino acid identity with mcr-3, the most closely related allele. In addition, it was most commonly found in Salmonella, whereas other mcr genes were more common in E. coli and Klebsiella. The first isolate identified with mcr-9 was susceptible to colistin, with an MIC of 0.25 to 0.5 μg/ml, although mcr-9 did confer colistin MICs of >2.5 μg/ml when cloned and overexpressed in the laboratory (15). Further work found that mcr-9 expression was inducible in the presence of colistin when located upstream of the two-component system qseBC (16). A subsequent study with 30 mcr-9-positive isolates from various Gram-negative organisms found that all were susceptible to colistin and lacked the qseBC regulatory operon (17). Additional studies reported conflicting results as to the phenotype associated with mcr-9 depending upon the isolate collection and species, indicating uncertainty regarding when this gene confers elevated colistin MICs (18, 19). The genomic structure of mcr-9-associated plasmids or chromosomes have not been substantially characterized, although there has been some association with IncHI plasmids (17, 20). To help determine whether mcr-9 conferred decreased susceptibility to colistin and may have clinical relevance, we characterized 100 isolates with mcr-9 recovered from the NARMS retail meat program by whole-genome sequencing (WGS) and colistin susceptibility testing.
RESULTS
Prevalence of mcr-9.
After mcr-9 was first identified, we sought to determine whether this gene was in any bacterial isolates collected as part of routine NARMS retail meat surveillance. Unlike previous mcr genes, most isolates identified in the NCBI Isolates Browser (1,052/1,862) were from Salmonella enterica. Upon additional analysis, we found that many of these S. enterica isolates (301 [28.6%]) were from NARMS retail meat sampling.
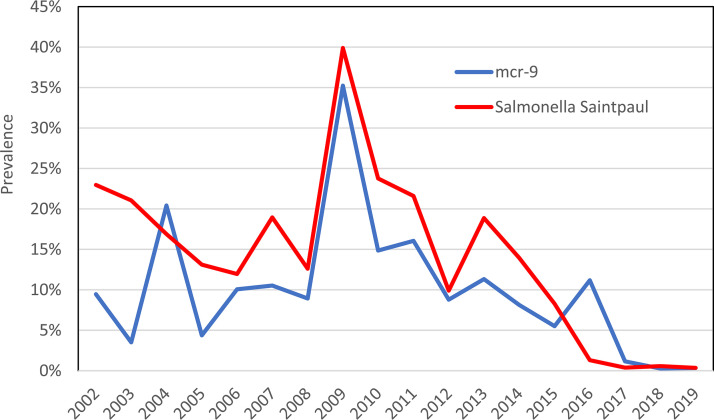
We further analyzed the assembled genomes and confirmed that each was indeed positive for mcr-9 by BLAST analysis. An additional six E. coli isolates from NARMS retail meat sampling also had mcr-9. Each isolate had mcr-9 with 100% identity and length to the reference sequence. NARMS performs sampling of retail beef, pork, turkey, and chicken products as part of its surveillance. However, 95.3% (287/301) of isolates with mcr-9 were from retail turkey products. Despite the recent identification of mcr-9, we found that its occurrence was not recent. In fact, it was found in all years of NARMS retail meat testing, from 2002 to 2019, with its prevalence among turkey isolates peaking at 35.2% in 2009 (Fig. 1). Further analysis showed the 301 mcr-9 isolates comprised 12 different serotypes, with Salmonella enterica serovar Saintpaul representing 75.1% of all isolates (Table 1). Therefore, the mcr-9 prevalence in ground turkey tracks closely with that of Salmonella Saintpaul (Fig. 1). All Saintpaul isolates were in the same single nucleotide polymorphism (SNP) cluster, PDS000004383.101, in the NCBI Isolates Browser, indicating that they were all within 50 SNPs of one another. For E. coli, the six isolates with mcr-9 were all from 2018 and 2019 retail sampling, with one isolate from ground turkey, four from retail chicken, and one from ground beef.
FIG 1.
mcr-9 prevalence among ground turkey isolates over time.
TABLE 1.
Salmonella enterica serotypes among NARMS retail isolates with mcr-9
| Serotype | No. of isolates in: |
|||
|---|---|---|---|---|
| Ground turkey | Retail chicken | Pork chop | Ground beef | |
| I 4,[5],12:i:- | 1 | 2 | ||
| Agona | 1 | |||
| Albany | 6 | |||
| Heidelberg | 30 | 1 | ||
| Johannesburg | 1 | |||
| Kentucky | 2 | 1 | ||
| Litchfield | 1 | |||
| Mbandaka | 2 | |||
| Montevideo | 1 | |||
| Saintpaul | 221 | 2 | 3 | |
| Schwarzengrund | 23 | |||
| Senftenberg | 2 | 1 | ||
| Total | 287 | 6 | 4 | 4 |
Additional analysis determined whether any isolates had the qseBC two-component system, which is thought to induce mcr-9 expression (16). We found these genes in 10 isolates, including 4 E. coli and 6 Salmonella isolates (see Table S1 in the supplemental material).
Susceptibility testing.
Broth microdilution testing was performed to determine the relationship between MIC and the presence of the mcr gene. We included a variety of different isolates, representing those isolated in each year from 2002 to 2019, and included those from all retail meat sources in testing (Table S1). A total of 105 isolates were tested, including 99 S. enterica and 6 E. coli isolates, and all were susceptible to colistin, with MICs of ≤1 μg/ml (Fig. 2). These results were true for all isolates tested, including all 10 isolates with qseBC (Table S1), which in some other studies had elevated colistin MICs. Furthermore, all isolates had MICs of ≤2 μg/ml for polymyxin B (Table S1), which had not previously been tested in other studies with mcr-9.
FIG 2.
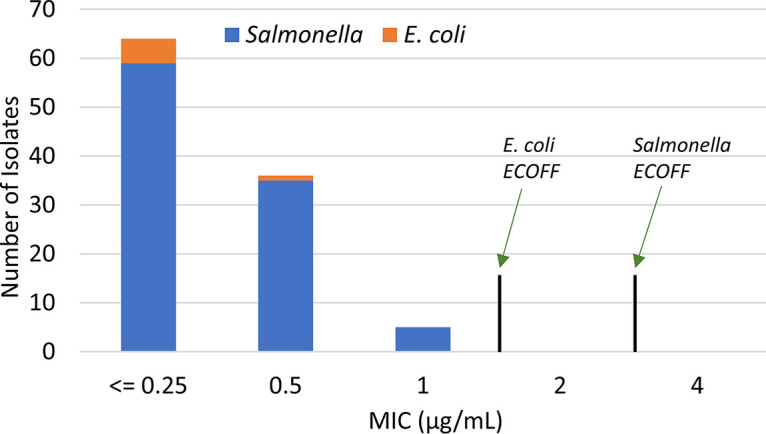
Colistin MICs of isolates with mcr-9.
Genomic structure of mcr-9-associated plasmids and chromosomes.
To further characterize the genetic context of the mcr-9 genes, we performed long-read sequencing using Pacific Biosciences technology. We sought to sequence diverse isolates to understand the various plasmids that carried this gene. Surprisingly, 6 of the 12 isolates had mcr-9 on the chromosome, with the remaining isolates having it on IncHI plasmids (Table S2).
The IncHI plasmids ranged from 269 to 340 kb, and all contained genes encoding mercury, tellurium, and copper resistance (Fig. S1 to S3). The presence of additional resistance genes varied among the plasmids; notably, the E. coli plasmid in isolate N18EC0432 possessed blaTEM-1, blaSHV-12, sul1, sul2, ereA, and aac(6′)-IIc (Fig. S2). These genes encode resistance to beta-lactams, sulfonamides, macrolides, and aminoglycosides. Of importance is blaSHV-12, an extended-spectrum-beta-lactamase gene whose presence could negatively impact treatment of associated infections. While most sequences did not have significant homology to known plasmids, the plasmid from isolate N53023 was highly related to an unknown plasmid in Enterobacter hormaechei (GenBank accession no. CP027144), which has mcr-9 and has public data in GenBank listing the colistin MIC as ≤0.25 μg/ml (Fig. S3). All chromosomal copies of mcr-9 appear to have been mobilized by integration of either entire or partial IncHI plasmids.
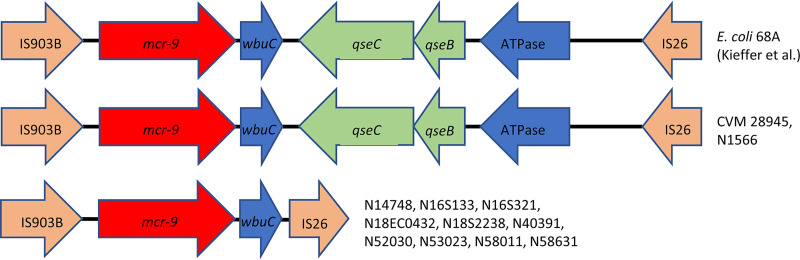
To better understand the genetic context of mcr-9 in our isolates, we compared the sequences from our closed genomes to that from the colistin-resistant isolate E. coli 68A (16). Two of the isolates had genetic structures identical to that of E. coli 68A, including the regulatory genes qseBC, but still had colistin MICs of ≤0.5 μg/ml (Fig. 3). There was only one SNP difference in our sequences compared to that of E. coli 68A in the entire 8.4-kb sequence, which was in the coding region of IS903B and unlikely to affect the function of mcr-9 or qseBC. The remaining 10 isolates with closed genomes had identical mcr-9 genetic structures to one another, and all lacked the qseBC genes.
FIG 3.
Genetic context of mcr-9 genes. The mcr-9 genes and their surrounding environments are shown. GenBank accession numbers for these are as follows: GCA_900500325 (E. coli 68A), CP048303 (CVM 28945), CP048299 (N1566), CP048926 (N14748), CP049986 (N16S133), CP049313 (N16S321), CP048293 (N18EC0432), CP049312 (N18S2238), CP049983 (N40391), CP049981 (N52030), CP049310 (N53023), CP049309 (N58011), and CP049307 (N58631).
Conjugations.
To further confirm the phenotype and transmissibility of plasmids associated with mcr-9, we performed conjugation assays on four different isolates. Conjugation was successful for one isolate, N53023, into the recipient strain DH5α, indicating that the IncHI plasmid is transmissible. Conjugations may have been more successful had they been conducted 26°C, as has been found optimal for IncHI plasmids (21). The transconjugant remained susceptible to colistin and polymyxin B (Table S1) but was resistant to other antimicrobials as predicted by the resistance genes encoded by the mcr-9 IncHI plasmid. These results confirm the prior susceptibility testing results that mcr-9 does not confer polymyxin resistance in our isolate collection. However, the initial isolate and transconjugant both had MICs of ≤0.25 μg/ml, so differences in MICs below the level of detection would not have been found based on our testing.
DISCUSSION
Microbial WGS has greatly expanded the data set of presumptive coding regions from which to infer function through gene relatedness surveys. The putative colistin resistance gene mcr-9 was first identified by whole-genome sequencing by its 64.5% amino acid identity to mcr-3. We examined our database of over 4,000 NARMS genomes and found mcr-9 in hundreds of isolates. This discovery was concerning since other mcr alleles had not previously been found in our isolate collection. However, our susceptibility testing and conjugation assays show that the presence of mcr-9 was not associated with resistance to colistin or polymyxin B.
With the rapidly growing body of genomic data, it is especially important to study the genetic context and test large numbers of isolates to make firm assertions about novel resistance alleles. By testing over 100 Salmonella and E. coli isolates with mcr-9 and closing the genomes of many of these, we have provided a basis to further understand differences in resistance phenotypes of bacteria with mcr-9.
Based on our results, it appears that mcr-9 is not as concerning as other mcr alleles, most of which consistently display colistin resistance. In the absence of a colistin resistance phenotype in our isolates, it is also unclear what function, if any, mcr-9 may have. However, it is worth noting that not all isolates with mcr genes have been found to be colistin resistant (22). These results also demonstrate the continued value of antimicrobial susceptibility testing in certain contexts, where conclusions resulting from whole-genome sequencing may not be clear. Although mcr-9 does not appear to be an emerging public health threat for now, changes in genetic context or the accumulation of mutations may affect its ability to impact colistin resistance. The closest reported allele to mcr-9 is mcr-3, and some bacteria with this allele are also colistin susceptible (23). It is important to note that all copies of mcr-9 in our collection had 100% identity and length to one another, so it is possible that changes in sequence could alter the effects of mcr-9 on colistin susceptibility. In addition, each isolate had mcr-9 on an IncHI plasmid or integrated into the chromosome from an IncHI plasmid, so a different genetic context could result in increased expression and a different resistance phenotype. These findings of association with IncHI plasmids are consistent with some previous findings (17, 20). Interestingly, the IncHI plasmids in our isolates were diverse both in overall genomic structure and in genomic context around the mcr-9 genes (Supplemental Fig. S1 to S3).
The two-component system qseBC was downstream of mcr-9 in several of our isolates, including two with closed genomes (Fig. 3). It is unclear why the presence of qseBC did not discernibly change the observed phenotypes, considering that the genetic context around mcr-9 was identical to that of E. coli 68A from Kieffer et al. (16). It is possible that the unusual O15:H6 serotype of E. coli 68A in some way contributed to the observed phenotype, as serotype-dependent colistin susceptibility has also been observed in Salmonella (24). Overexpression of mcr genes in E. coli TOP10 showed increased MICs from 0.03 μg/ml to 0.15 μg/ml for mcr-9, compared to 4 μg/ml for mcr-1 (16). We did not perform induction experiments, but preinduction of mcr-9 in the presence of colistin is not required to accurately measure MICs by broth microdilution (16). These results indicate that the effectiveness of qseBC in inducing mcr-9 expression may be context dependent and differ among isolates with different strain backgrounds.
Interestingly, since mcr-9 is frequently present on mobile genetic elements with other resistance genes, it means that the use of these other antimicrobials has the potential to coselect for the continued presence of mcr-9. In addition, various metal resistance genes, encoding resistance to silver, mercury, arsenic, copper, tellurium, and others, are present on these plasmids, providing another potential avenue for coselection for mcr-9.
NARMS will continue to detect colistin resistance genes as part of routine whole-genome sequencing and recently added colistin as part of routine antimicrobial susceptibility testing. Thus, ongoing work will determine whether colistin resistance emerges among bacteria collected as part of NARMS sampling and, if so, what the resulting mechanisms are.
MATERIALS AND METHODS
Data mining.
Isolates with mcr-9 were identified using the NCBI Pathogen Detection Isolates Browser, which annotates antimicrobial resistance genes using AMRFinderPlus (25). Initial results included investigation of all sequences with mcr-9 among Salmonella enterica isolates present in the browser, among a total of more than 200,000 total Salmonella enterica genomes. Further analysis focused on NARMS isolates in BioProject PRJNA292661, which contains sequencing data from more than 4,000 isolates. Search results were collected on 13 December 2019.
Colistin susceptibility testing.
Isolates with mcr-9 were tested for susceptibility to colistin and polymyxin B using standard broth microdilution techniques, following CLSI standards (36). The panels GNX3F from Trek Diagnostics were used, which have colistin and polymyxin B concentrations of 0.25 μg/ml to 4 μg/ml. EUCAST epidemiological cutoffs (ECOFFs) and provisional ECOFFs were used to identify isolates with elevated MICs (26). Quality control strains E. coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used, along with a colistin-resistant WHO reference strain with mcr-4 (Salmonella Kedougou WHO-S18-2) (27).
Whole-genome sequencing and analysis.
Whole-genome sequencing was performed by short-read sequencing on the Illumina MiSeq using v3 chemistry, as previously described (28). Twelve isolates were selected for long-read sequencing, with DNA libraries prepared using a 10-kb template preparation protocol with SMRTbell template prep kit v1.0. Sequencing was performed using Pacific Biosciences technology on the Sequel platform with sequencing kit 3.0, as previously described (29). The long reads were assembled to contigs by HGAP4, and contigs were circularized by Circlator (30, 31). Assembled genomes were then polished with Illumina reads by Pilon (32). Even and continuous coverage was assessed to determine that there were no misassemblies. Genes were annotated by Prokka in the depicted figures (33), and BLASTn was used to compare plasmid sequences with each other and with those in GenBank. PlasmidFinder was used to identify the plasmid types (34).
Conjugation.
Conjugation assays were conducted as previously described (35). Briefly, recipient DH5α E. coli cells were resistant to nalidixic acid, with donor N53023 resistant to gentamicin, as encoded by aac(3)-VIa on the mcr-9 IncHI plasmid. Mating occurred on blood agar plates at 37°C for 16 h. The conjugated cells were resuspended in 500 μl of LB broth, and 50 μl of the resulting mixture was plated onto doubly selective agars (nalidixic acid plus gentamicin). Individual isolates were confirmed not to grow on these doubly selective agars. Any resulting colonies were screened by whole-genome sequencing for recipient genomes and the presence of mcr-9 plasmid sequence. Subsequent antimicrobial susceptibility testing was performed to identify any changes in colistin resistance phenotypes.
Accession number(s).
Whole-genome sequencing data were submitted to NCBI with accession numbers for short-read sequences in Table S1 and long-read sequences in Table S2.
Supplementary Material
ACKNOWLEDGMENTS
The views expressed in this article are those of the authors and do not necessarily reflect the official policy of the Department of Health and Human Services, the U.S. Food and Drug Administration, or the U.S. government. Reference to any commercial materials, equipment, or process does not in any way constitute approval, endorsement, or recommendation by the Food and Drug Administration.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Lim LM, Ly N, Anderson D, Yang JC, Macander L, Jarkowski A III, Forrest A, Bulitta JB, Tsuji BT. 2010. Resurgence of colistin: a review of resistance, toxicity, pharmacodynamics, and dosing. Pharmacotherapy 30:1279–1291. doi: 10.1592/phco.30.12.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupta S, Govil D, Kakar PN, Prakash O, Arora D, Das S, Govil P, Malhotra A. 2009. Colistin and polymyxin B: a re-emergence. Indian J Crit Care Med 13:49–53. doi: 10.4103/0972-5229.56048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rhouma M, Beaudry F, Theriault W, Letellier A. 2016. Colistin in pig production: chemistry, mechanism of antibacterial action, microbial resistance emergence, and One Health perspectives. Front Microbiol 7:1789. doi: 10.3389/fmicb.2016.01789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anonymous. 2016. European Medicines Agency to review guidance on colistin use in animals. Vet Rec 178:55. [DOI] [PubMed] [Google Scholar]
- 5.Walsh TR, Wu Y. 2016. China bans colistin as a feed additive for animals. Lancet Infect Dis 16:1102–1103. doi: 10.1016/S1473-3099(16)30329-2. [DOI] [PubMed] [Google Scholar]
- 6.Bialvaei AZ, Samadi Kafil H. 2015. Colistin, mechanisms and prevalence of resistance. Curr Med Res Opin 31:707–721. doi: 10.1185/03007995.2015.1018989. [DOI] [PubMed] [Google Scholar]
- 7.Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 8.Cannatelli A, Giani T, Antonelli A, Principe L, Luzzaro F, Rossolini GM. 2016. First detection of the mcr-1 colistin resistance gene in Escherichia coli in Italy. Antimicrob Agents Chemother 60:3257–3258. doi: 10.1128/AAC.00246-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiou CS, Chen YT, Wang YW, Liu YY, Kuo HC, Tu YH, Lin AC, Liao YS, Hong YP. 2017. Dissemination of mcr-1-carrying plasmids among colistin-resistant Salmonella strains from humans and food-producing animals in Taiwan. Antimicrob Agents Chemother 61:e00338-17. doi: 10.1128/AAC.00338-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Pilato V, Arena F, Tascini C, Cannatelli A, Henrici De Angelis L, Fortunato S, Giani T, Menichetti F, Rossolini GM. 2016. mcr-1.2, a new mcr variant carried on a transferable plasmid from a colistin-resistant KPC carbapenemase-producing Klebsiella pneumoniae strain of sequence type 512. Antimicrob Agents Chemother 60:5612–5615. doi: 10.1128/AAC.01075-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borowiak M, Fischer J, Hammerl JA, Hendriksen RS, Szabo I, Malorny B. 2017. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J Antimicrob Chemother 72:3317–3324. doi: 10.1093/jac/dkx327. [DOI] [PubMed] [Google Scholar]
- 12.Yin W, Li H, Shen Y, Liu Z, Wang S, Shen Z, Zhang R, Walsh TR, Shen J, Wang Y. 2017. Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. mBio 8:e00543-17. doi: 10.1128/mBio.00543-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henig O, Rojas LJ, Bachman MA, Rudin SD, Brennan BM, Soehnlen MK, Jones KL, Mills JP, Dombecki CR, Valyko AM, Marshall SH, Bonomo RA, Kaye KS, Washer L. 2019. Identification of four patients with colistin-resistant Escherichia coli containing the mobile colistin resistance mcr-1 gene from a single health system in Michigan. Infect Control Hosp Epidemiol 40:1059–1062. doi: 10.1017/ice.2019.177. [DOI] [PubMed] [Google Scholar]
- 14.Monte DF, Nelson V, Cerdeira L, Keelara S, Greene S, Griffin D, Rath S, Hall R, Page N, Fedorka-Cray PJ, Thakur S. 2019. Multidrug- and colistin-resistant Salmonella enterica 4,[5],12:i:- sequence type 34 carrying the mcr-3.1 gene on the IncHI2 plasmid recovered from a human. J Med Microbiol 68:986–990. doi: 10.1099/jmm.0.001012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carroll LM, Gaballa A, Guldimann C, Sullivan G, Henderson LO, Wiedmann M. 2019. Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible Salmonella enterica serotype Typhimurium isolate. mBio 10:e00853-19. doi: 10.1128/mBio.00853-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kieffer N, Royer G, Decousser JW, Bourrel AS, Palmieri M, Ortiz De La Rosa JM, Jacquier H, Denamur E, Nordmann P, Poirel L. 2019. mcr-9, an inducible gene encoding an acquired phosphoethanolamine transferase in Escherichia coli, and its origin. Antimicrob Agents Chemother 63:e00965-19. doi: 10.1128/AAC.00965-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borjesson S, Greko C, Myrenas M, Landen A, Nilsson O, Pedersen K. 2020. A link between the newly described colistin resistance gene mcr-9 and clinical Enterobacteriaceae isolates carrying blaSHV-12 from horses in Sweden. J Glob Antimicrob Resist 20:285–289. doi: 10.1016/j.jgar.2019.08.007. [DOI] [PubMed] [Google Scholar]
- 18.Chavda KD, Westblade LF, Satlin MJ, Hemmert AC, Castanheira M, Jenkins SG, Chen L, Kreiswirth BN. 2019. First report of blaVIM-4- and mcr-9-coharboring Enterobacter species isolated from a pediatric patient. mSphere 4:e00629-19. doi: 10.1128/mSphere.00629-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan Y, Li Y, Wang G, Li C, Xiang L, She J, Yang Y, Zhong F, Zhang L. 2019. Coproduction of MCR-9 and NDM-1 by colistin-resistant Enterobacter hormaechei isolated from bloodstream infection. Infect Drug Resist 12:2979–2985. doi: 10.2147/IDR.S217168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borowiak M, Baumann B, Fischer J, Thomas K, Deneke C, Hammerl JA, Szabo I, Malorny B. 2020. Development of a novel mcr-6 to mcr-9 multiplex PCR and assessment of mcr-1 to mcr-9 occurrence in colistin-resistant Salmonella enterica isolates from environment, feed, animals and food (2011–2018) in Germany. Front Microbiol 11:80. doi: 10.3389/fmicb.2020.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor DE, Levine JG. 1980. Studies of temperature-sensitive transfer and maintenance of H incompatibility group plasmids. J Gen Microbiol 116:475–484. doi: 10.1099/00221287-116-2-475. [DOI] [PubMed] [Google Scholar]
- 22.Li H, Yang L, Liu Z, Yin W, Liu D, Shen Y, Walsh T, Shao B, Wang Y. 2018. Molecular insights into functional differences between mcr-3- and mcr-1-mediated colistin resistance. Antimicrob Agents Chemother 62:e00366-18. doi: 10.1128/AAC.00366-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen Y, Xu C, Sun Q, Schwarz S, Ou Y, Yang L, Huang Z, Eichhorn I, Walsh TR, Wang Y, Zhang R, Shen J. 2018. Prevalence and genetic analysis of mcr-3-positive Aeromonas species from humans, retail meat, and environmental water samples. Antimicrob Agents Chemother 62:e00404-18. doi: 10.1128/AAC.00404-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ricci V, Zhang D, Teale C, Piddock L. 2020. The O-antigen epitope governs susceptibility to colistin in Salmonella enterica. mBio 11:e02831-19. doi: 10.1128/mBio.02831-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feldgarden M, Brover V, Haft DH, Prasad AB, Slotta DJ, Tolstoy I, Tyson GH, Zhao S, Hsu CH, McDermott PF, Tadesse DA, Morales C, Simmons M, Tillman G, Wasilenko J, Folster JP, Klimke W. 2019. Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob Agents Chemother 63:e00483-19. doi: 10.1128/AAC.00483-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tyson GH, Bodeis-Jones S, Caidi H, Cook K, Dessai U, Haro J, McCullough AE, Meng J, Morales CA, Lawrence JP, Tillman GE, Winslow A, Miller RA. 2018. Proposed epidemiological cutoff values for ceftriaxone, cefepime, and colistin in Salmonella. Foodborne Pathog Dis 15:701–704. doi: 10.1089/fpd.2018.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rebelo AR, Bortolaia V, Kjeldgaard JS, Pedersen SK, Leekitcharoenphon P, Hansen IM, Guerra B, Malorny B, Borowiak M, Hammerl JA, Battisti A, Franco A, Alba P, Perrin-Guyomard A, Granier SA, De Frutos Escobar C, Malhotra-Kumar S, Villa L, Carattoli A, Hendriksen RS. 2018. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Euro Surveill 23:17-00672. doi: 10.2807/1560-7917.ES.2018.23.6.17-00672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDermott PF, Tyson GH, Kabera C, Chen Y, Li C, Folster JP, Ayers SL, Lam C, Tate HP, Zhao S. 2016. Whole-genome sequencing for detecting antimicrobial resistance in nontyphoidal Salmonella. Antimicrob Agents Chemother 60:5515–5520. doi: 10.1128/AAC.01030-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tyson GH, Li C, Ceric O, Reimschuessel R, Cole S, Peak L, Rankin SC. 2019. Complete genome sequence of a carbapenem-resistant Escherichia coli isolate with blaNDM-5 from a dog in the United States. Microbiol Resour Announc 8:e00872-19. doi: 10.1128/MRA.00872-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 31.Hunt M, Silva ND, Otto TD, Parkhill J, Keane JA, Harris SR. 2015. Circlator: automated circularization of genome assemblies using long sequencing reads. Genome Biol 16:294. doi: 10.1186/s13059-015-0849-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 34.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, Møller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tyson GH, Sabo JL, Hoffmann M, Hsu CH, Mukherjee S, Hernandez J, Tillman G, Wasilenko JL, Haro J, Simmons M, Wilson Egbe W, White PL, Dessai U, McDermott PF. 2018. Novel linezolid resistance plasmids in Enterococcus from food animals in the USA. J Antimicrob Chemother 73:3254–3258. doi: 10.1093/jac/dky369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clinical and Laboratory Standards Institute. 2020. Performance standards for antimicrobial susceptibility testing, M100, 30th ed. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.