FIG 2.
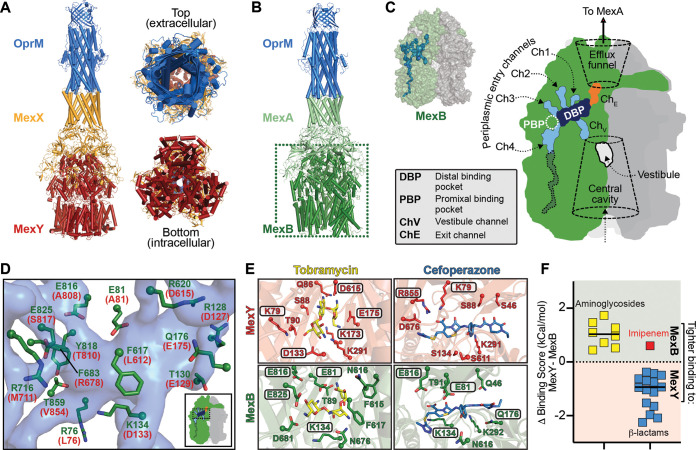
Substrate selectivity by MexB and MexY is determined by a Goldilocks binding affinity. (A) Three views of the computational model of the P. aeruginosa MexXY-OprM efflux pump. (B) Cryo-electron microscopy (cryo-EM) structure of the P. aeruginosa MexAB-OprM efflux pump (PDB code 6IOL) used to guide modeling of MexXY-OprM. (C) Surface representation of the MexB trimer with a single protomer colored green (other protomers are light and dark gray), with internal channels and pockets determined using Caver shown as a blue surface (this calculation was performed on the protomer in the binding state). A schematic of the same MexB trimer indicating individual channels/pockets, etc., is also shown. (D) Zoomed view of residues lining part of the DBP (semitransparent blue surface) in MexB (green) or the MexY model (red). (E) The aminoglycoside tobramycin and β-lactam cefoperazone can be docked in similar locations within the DBPs of MexY or MexB. Boxed residues making interactions with each drug are those predicted by the evolutionary analyses to be potentially critical for substrate discrimination by these efflux systems. (F) Differences of docking scores for each transporter (MexY-MexB [Table 2]) are plotted for each antibiotic class (aminoglycosides and β-lactams). Note that, consistent with our findings, imipenem (red) is not an efflux substrate of MexAB-OprM.