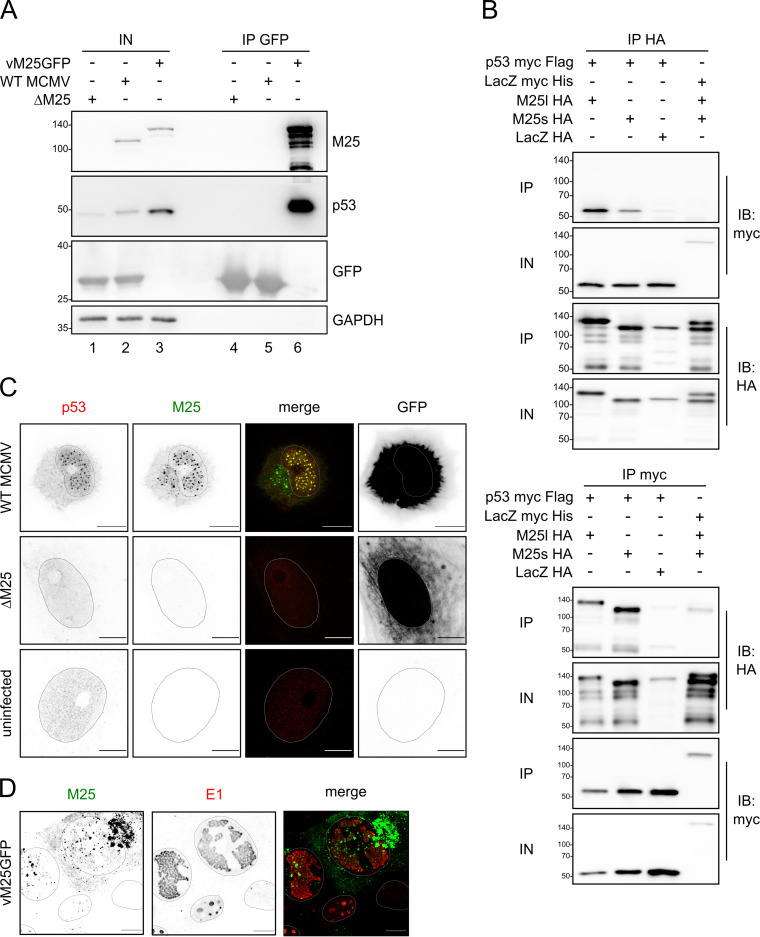
FIG 2.
M25 proteins interact with p53. (A) NIH 3T3 cells were infected with the indicated viruses (at MOI 1) and harvested 36 h p.i. GFP and GFP-tagged M25 proteins were precipitated with GFP nanobodies, and immunoblotting was performed with antibodies for the specified proteins. (B) p53-deficient HCT116 cells were transfected with plasmids coding for the indicated proteins, and immunoprecipitation was performed with an HA (top panel) or a myc antibody (bottom panel) using cell lysates prepared 24 h posttransfection. Antibodies used for immunoblotting are indicated in the right margin. IP, precipitated proteins; IN, input lysates. (C) MEF were infected with WT MCMV or the ΔM25 mutant at an MOI of 1 or were left uninfected, and at 36 h p.i. the cells were labeled with the indicated antibodies and analyzed by confocal microscopy. Images to the left are depicted in inverse mode, with positive staining appearing in black and gray. Infected cells were identified based on GFP expression (right panels). Please note that the apparently different GFP distribution is due to the different effects of WT MCMV and the ΔM25 mutant on cell morphology. While WT MCMV-infected cells become spherical, leaving a small cytoplasmic rim around the cell nucleus, ΔM25-infected cells retain an elongated shape. (D) NIH 3T3 cells were infected with the tagged virus vM25GFP for 24 h, followed by immunolabeling with an antibody against the E1 protein. The M25-GFP fusion protein was detected via GFP fluorescence. Shapes of the nuclei were determined based on Hoechst staining. Images are representative of at least 20 cells analyzed per condition. Scale bars, 10 μm.