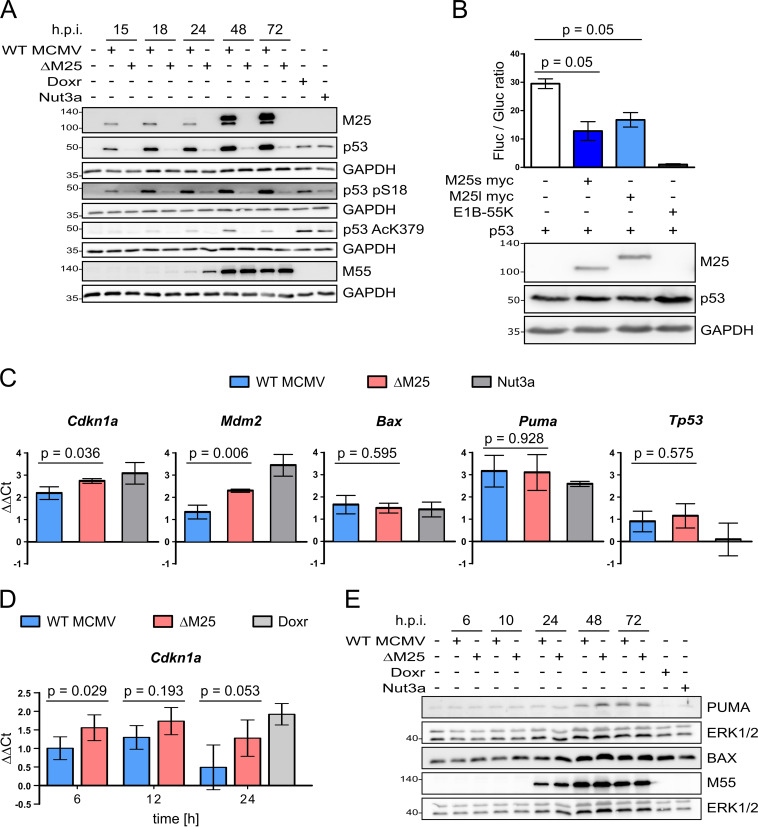
FIG 5.
Influence of M25 proteins on posttranslational modification of p53 and its transcriptional activity. (A) NIH 3T3 cells were either left uninfected or were infected with WT MCMV or the ΔM25 mutant (MOI of 3) or were treated with doxorubicin (Doxr; 0.5 μM for 24 h) or with Nutlin3a (Nut3a; 40 μM for 6 h). Cell lysates prepared at the indicated time points postinfection were analyzed by immunoblotting with antibodies for the specified proteins and the modified p53 versions. (B) p53-deficient HCT116 cells were transfected with the luciferase reporter plasmid PG13-luc, a Gaussia luciferase control plasmid and expression plasmids encoding p53, pM25s, pM25l, or the adenovirus E1B-55K protein as indicated. The luciferase activities were measured 48 h after transfection. The graph shows luciferase activities ± the standard deviations (SD) in cell lysates (ratios of firefly and Gaussia luciferase activity) of three biological replicates. A Mann-Whitney test (one-tailed) was used to analyze statistical significance. (C) Total RNA was isolated from NIH 3T3 cells infected with WT MCMV or ΔM25 (MOI of 3) for 36 h and, for comparison, from uninfected cells (either untreated or treated with Nutlin3a [5 μM]) and was subjected to qRT PCR analysis using gene-specific primers. (D) Total RNA was isolated from primary MEF infected with either WT MCMV or ΔM25 (MOI of 3) for the depicted duration and from cells that remained uninfected or were treated with doxorubicin (Doxr) for 24 h. qRT PCR was performed using Cdkn1a-specific primers. (C and D) The graphs depict the ΔΔCT values of the qRT-PCRs (log2-transformed values of expression levels in relation to the ones of uninfected cells). The means ± the SD of three to five biological replicates are shown. A t test (two-tailed) was used for statistical analysis. (E) NIH 3T3 cells were infected or treated as indicated in panel A, and cell lysates prepared at the indicated time points postinfection were analyzed by immunoblotting with antibodies specific for PUMA or BAX.