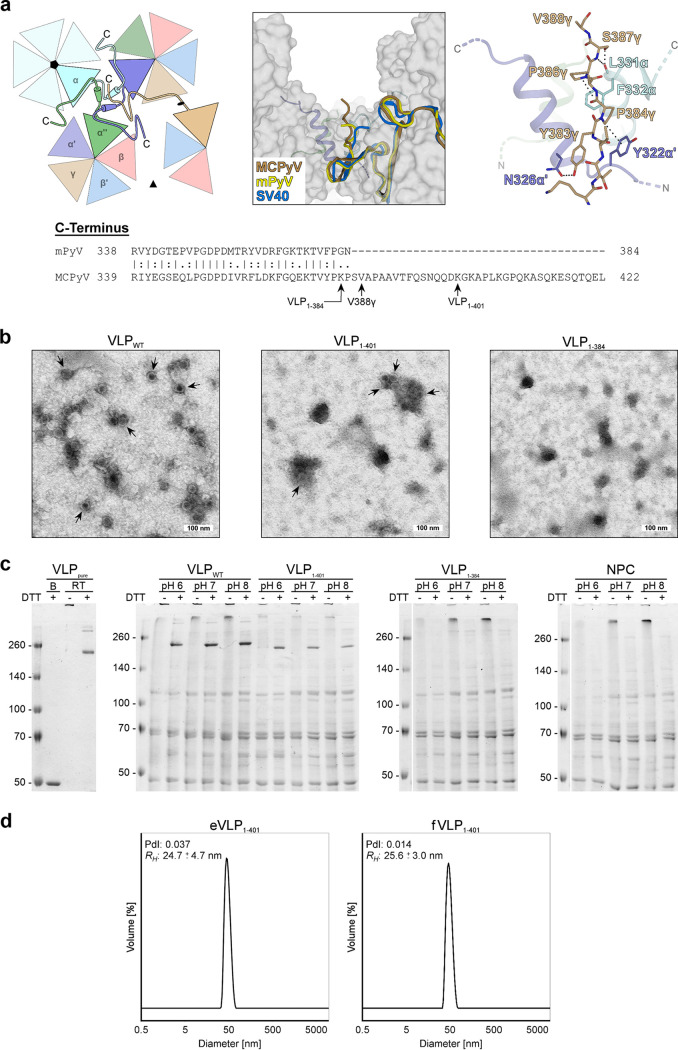
FIG 3.
The VP1 C terminus is essential for MCPyV VP1 integrity. (a) Sequence alignment of the MCPyV and mPyV VP1 C termini and conformation of the MCPyV C terminus exemplified by the γ-monomer. The C termini of MCPyV, mPyV, and SV40 γ-monomers are located at the three-helix bundle of the α-α′-α″ interface around the icosahedral 5-fold axis (left schematic representation) and point to the recessed regions between capsomers (middle). The conformations are similar, but the structure of MCPyV lacks 35 C-terminal amino acids (388 aa of 423 aa) after the last visible stretch shown here, in contrast to mPyV (362 aa of 362 aa) and SV40 (383 aa of 384 aa). The last visible residues (Lys380γ-Val388γ) of MCPyV are stabilized by hydrogen bonds and CH-π interactions (right). Stop codon positions of the MCPyV VLP mutants and the last visible residue (Val388γ) of the MCPyV VLP crystal structure are indicated. (b) Negative-stain transmission electron micrographs of C-terminally shortened mutants of MCPyV VLPs (VLP1–401 and VLP1–384) in comparison with the wild type (VLPWT). Shown are cell lysates after the initial high-salt precipitation step. Arrows indicate capsid assembly. (c) Comparison of MCPyV wild-type (VLPWT) and truncated (VLP1–401 and VLP1–384) VP1 isolates at different pHs via nonreducing SDS-PAGE. DTT treatment of the lysates at room temperature leads to a partial decay of VLPs to pentamers (∼250 kDa). In contrast to completely denatured MCPyV monomers (B) (lane 1), capsids of untreated samples remain intact and do not enter the gel. Purified VLPs (VLPpure) and lysates of untransfected 293 TT cells (NPC) were used as positive and negative controls. (d) Size distribution of purified VLP1–401 derived from dynamic light scattering. The polydispersity index (PdI) of the samples and the calculated hydrodynamic radius (RH) are indicated.