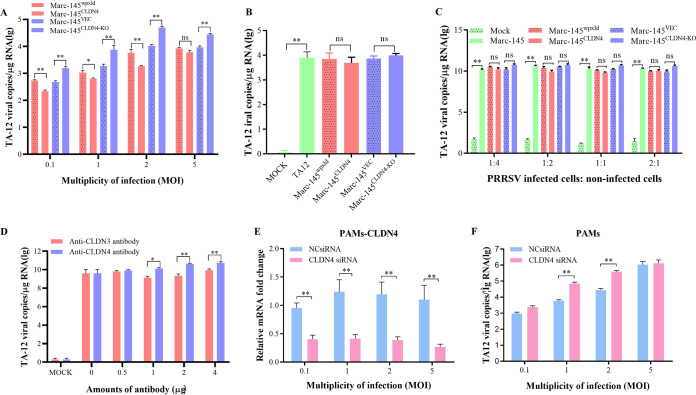
FIG 4.
CLDN4 could block PRRSV absorption. (A) Confirmation of the role of CLDN4 in absorption in MARC-145 cells. MARC-145CLDN4-KO and MARC-145CLDN4 cells and their corresponding control cell lines were infected with TA-12 at MOI of 0.1, 1, 2, and 5 and absorbed at 4°C for 1 h. After a washing with cold PBS, the cells were collected and the viral RNA was extracted. The attached PRRSV particles were evaluated by qPCR. (B) Confirmation of the role of CLDN4 in viral internalization. MARC-145CLDN4-KO and MARC-145CLDN4 cells and their corresponding control cell lines were infected with TA-12 at an MOI of 1, absorbed at 4°C for 1 h, and then shifted to 37°C for 4 h to allow internalization of the attached viruses. After a washing with low-pH PBS buffer, the cells were collected and the viral RNAs were extracted. The internalized PRRSV particles were evaluated by qPCR. (C) Confirmation of the role of CLDN4 in cell-to-cell transmission. Normal MARC-145 cells were infected with TA-12 at an MOI of 1 and collected at 24 h postinfection. After extensive washing in a low-pH buffer, they were mixed with MARC-145CLDN4-KO and MARC-145CLDN4 cells and their corresponding control cell lines at ratios of 1:4, 1:2, 1:1, and 2:1 before incubation for another 24 h. The cells were then collected, and the viral RNA was extracted. The PRRSV genome was evaluated by qPCR. (D) The blocking effects of an anti-CLDN4 antibody on PRRSV absorption. Anti-CLDN4 antibodies were incubated onto a monolayer of MARC-145 cells at 37°C for 1 h. After being washed, the cells were inoculated with TA-12 at an MOI of 0.1 and incubated at 37°C for 24 h. The cells were collected, and the viral RNAs were extracted. The genomic PRRSV was evaluated by qPCR. The anti-CLDN3 antibody was used as a negative control. The role of CLDN4 in absorption was confirmed in PAMs. PAMs were transfected with CLDN4 siRNAs for 24 h and incubated with TA-12 at MOI of 0.1, 1, 2, and 5 for 2 h at 4°C. After a washing with cold PBS, the cells were collected, and the total RNA was extracted. The CLDN4 knockdown was verified by qPCR (E); The PRRSV genome was evaluated by qPCR (F). All data are means and standard deviations (error bars) for three samples per group (*, P < 0.05; **, P < 0.01; ns, no significance). GAPDH was used as an internal control. All experiments were repeated at least three times with consistent results.