Members of the Pegivirus genus, family Flaviviridae, widely infect humans and other mammals, but are described as causing persistent infection and lacking pathogenicity. The efficiency of in vitro replication systems for pegivirus is poor, thus limiting investigation into viral replication steps. Because of that, the pathogenesis, cellular tropism, route of transmission, biology, and epidemiology of pegiviruses remain largely uncovered. Here, we report a phylogenetically distinct goose pegivirus (GPgV) that should be classified as a new species. GPgV proliferated in cell culture in a species- and cell-type-specific manner. Animal experiments show GPgV lymphotropism and promote goose parvovirus clinical manifestations. This study provides the first cell culture model for pegivirus, opening new possibilities for studies of pegivirus molecular biology. More importantly, our findings stand in contrast to the lack of identified pathogenicity of previously reported pegiviruses, which sheds lights on the pathobiology of pegivirus.
KEYWORDS: goose, pegivirus
ABSTRACT
Members of the Pegivirus genus, family Flaviviridae, widely infect humans and other mammals, including nonhuman primates, bats, horses, pigs, and rodents, but are not associated with disease. Here, we report a new, genetically distinct pegivirus in goose (Anser cygnoides), the first identified in a nonmammalian host species. Goose pegivirus (GPgV) can be propagated in goslings, embryonated goose eggs, and primary goose embryo fibroblasts, and is thus the first pegivirus that can be efficiently cultured in vitro. Experimental infection of GPgV in goslings via intravenous injection revealed robust replication and high lymphotropism. Analysis of the tissue tropism of GPgV revealed that the spleen and thymus were the organs bearing the highest viral loads. Importantly, GPgV could promote clinical manifestations of goose parvovirus infection, including reduced weight gain and 7% mortality. This finding contrasts with the lack of pathogenicity that is characteristic of previously reported pegiviruses.
IMPORTANCE Members of the Pegivirus genus, family Flaviviridae, widely infect humans and other mammals, but are described as causing persistent infection and lacking pathogenicity. The efficiency of in vitro replication systems for pegivirus is poor, thus limiting investigation into viral replication steps. Because of that, the pathogenesis, cellular tropism, route of transmission, biology, and epidemiology of pegiviruses remain largely uncovered. Here, we report a phylogenetically distinct goose pegivirus (GPgV) that should be classified as a new species. GPgV proliferated in cell culture in a species- and cell-type-specific manner. Animal experiments show GPgV lymphotropism and promote goose parvovirus clinical manifestations. This study provides the first cell culture model for pegivirus, opening new possibilities for studies of pegivirus molecular biology. More importantly, our findings stand in contrast to the lack of identified pathogenicity of previously reported pegiviruses, which sheds lights on the pathobiology of pegivirus.
INTRODUCTION
The family Flaviviridae is currently divided into four genera: Flavivirus, Hepacivirus, Pestivirus, and Pegivirus (1). The first three genera include a number of viruses that cause human and/or animal disease with high morbidity and mortality; well-known examples are yellow fever virus, dengue virus, West Nile virus, classical swine fever virus, and hepatitis C virus (HCV). In the Pegivirus genus, 11 species (Pegivirus A to K) have been assigned which infect humans and other mammals. The origins of human pegiviruses (HPgV-1, Pegivirus C; HPgV-2, Pegivirus H) are unknown, and there is no evidence indicating that any animal serves as a natural reservoir of these viruses. All known pegiviruses possess genomes characterized by the absence of an identified core protein coding region and variable arrangements of putative envelope genes. All pegiviruses possess internal ribosomal entry sites (IRESs); human pegivirus type 2 (HPgV-2) and other group 2 pegiviruses have IRESs structurally similar to the type IV IRES of hepaciviruses (2), while other pegiviruses, such those of HPgV-1 and other group 1 pegiviruses, are structurally distinct.
All pegiviruses have the ability to cause persistent infection in their respective hosts (3). Currently, pegiviruses are described as lacking any confirmed pathogenicity. Because of the lack of a suitable in vitro culture system, the association between acute hepatitis in horses (Theiler’s disease) and Theiler’s disease-associated pegivirus (TDAV, also known as horse origin Pegivirus D) remains inferential (4). In contrast, it has been suggested that HPgV-1 infection improves the outcome of human immunodeficiency virus (HIV-1) infections (2, 5, 6). Understanding the pathogenic potential of pegiviruses and their ability to modify symptoms caused by other viruses is therefore necessary. This is, however, very challenging due to the lack of an “efficient” cell culture system for pegivirus, which limits investigation into viral replication steps. Because of this, the pathogenesis, cellular tropism, route of transmission, biology, and epidemiology of pegiviruses remain largely unknown.
Here, we report the discovery of the first nonmammalian pegivirus in Southwest China. The virus was named goose pegivirus (GPgV) to reflect its host, in which it reduces weight gain and shows high lymphotropism. Unlike any known pegivirus, GPgV replicates well in cell culture; however, in vitro its host cell range was limited to goose embryo fibroblasts (GEFs). Experimental in vivo inoculation of GPgV was used to reveal the tissue tropism and to determine viral loads in different organs. The outcome of coinfection of GPgV and goose parvovirus (GPV) was also investigated both in vivo and in vitro. Our data has provided the first substantive experimental information on the biology and pathogenesis of a member of the Pegivirus genus.
RESULTS
Sample collection.
In past decades, several goose farms in Sichuan Province and the Chongqing municipality of China have reported sporadic goose disease of unknown etiology and high morbidity and mortality. The main clinical signs associated with naturally occurring disease are decreased feed efficiency, uneven growth, loss of back down, and watery diarrhea. The duodenum mucosa demonstrates marked hyperemia, and there is rectal distension with white and yellowish discharge. Disease is most frequently observed in goslings during the first 1 to 3 weeks of life. The sick goslings usually show severe diarrhea and sudden death; in most cases, no other obvious clinical symptoms are observed. In an attempt to reveal the causative agent of the disease, the duodenum, rectum, liver, kidney, spleen, and brain from dead geese were sampled and analyzed. No infection with known bacteria was revealed. PCR, reverse transcriptase PCR (RT-PCR), and nested RT-PCR analysis revealed that the samples were also negative for known viruses, including GPV, goose circovirus, goose astrovirus, goose coronaviruses, goose paramyxovirus, and avian influenza virus.
GPgV genome sequencing and assembly.
To identify the possible pathogen causing the disease, material isolated from intestine and liver homogenates of dead geese was used to infect 9-day-old embryonated goose eggs through allantoic cavity injection. The putative pathogen was passaged 10 times, after which DNA and RNA were extracted from allantoic fluid and analyzed using unbiased metagenomic next-generation sequencing (NGS). No sequences of known DNA viruses were found. For the RNA sample, a total of 6.79 million raw reads were generated. These sequences were assembled using Trinity software, resulting in 202 unigenes; five of them were matched to pegivirus by the BLASTx NCBI database and assembled into two long sequences. RNA sequence 1 (GPgV-1) was 11,479 nucleotides (nt) in length and encoded a protein that exhibited 42% amino acid identity and 71% coverage with TDAV, while sequence 2 was 11,195 nt in length and the corresponding protein exhibited 39% amino acid identity and 67% coverage with TDAV. To verify these results and obtain a complete sequence of GPgV, five DNA segments covering the entire genome were PCR amplified, cloned, and sequenced using Sanger sequencing (the primers used are shown in Fig. 1 and listed in Table 1). Only fragments corresponding to GPgV-1 were obtained and analyzed. For unknown reasons, RT-PCR, as well as nested PCR, failed to amplify fragments corresponding to RNA sequence 2 (GPgV-2).
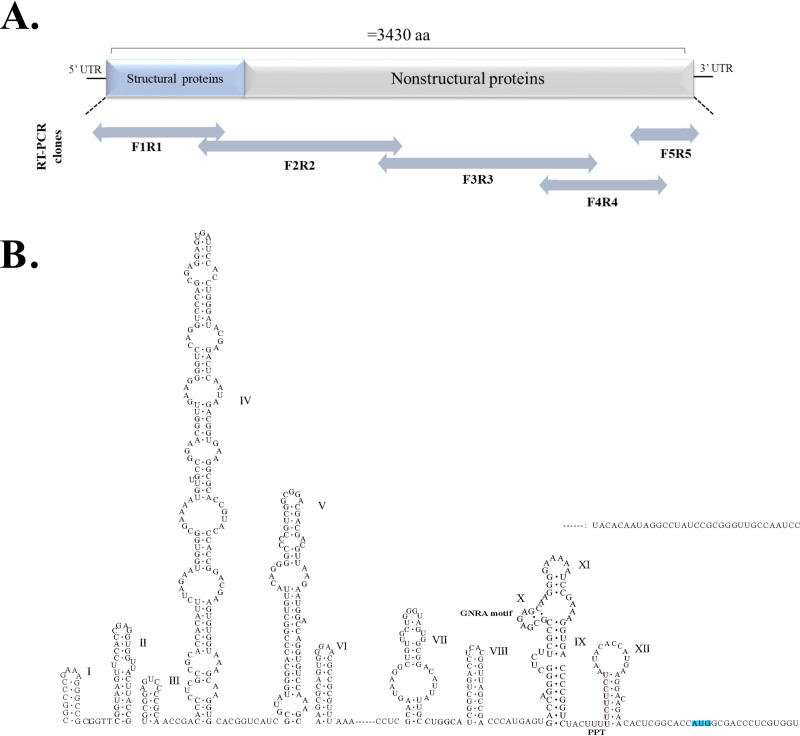
FIG 1.
Schematic presentation of the GpgV genome and location of PCR fragments used for its sequencing and predicted RNA secondary structures in the 5′ region of GPgV genome. (A) Two-way arrows indicate the relative sizes and locations of the overlapping PCR fragments. (B) Sequence corresponding to the 5′ UTR and 14 nt located downstream of the translation initiation codon (indicated in blue) was analyzed using Mfold. IRES-associated sequences (GNRA motif and PPT) are indicated.
TABLE 1.
Primers used in this study
| Primer | Purpose | Source or reference | Sequence (5′–3′) | Location in genome (nt)a |
|---|---|---|---|---|
| GPgV F1 | To confirm the genome | This study | GACATTATTGCCTGGTATCG | 416-435 |
| GPgV R1 | To confirm the genome | This study | CACAGCCACGAAAGCATC | 2192-2209 |
| GPgV F2 | To confirm the genome | This study | CAAGCACGAATTGGAAACA | 1892-1910 |
| GPgV NF2 | Nested RT-PCR | This study | GGTCTACGCCTACAACCACT | 3751-3770 |
| GPgV R2 | To confirm the genome; nested RT-PCR | This study | TTCTCCTCAATGGCTGTGC | 5286-5304 |
| GPgV F3 | To confirm the genome | This study | TCTCATCTTCTGCCACTCAAA | 5092-5112 |
| GPgV R3 | To confirm the genome | This study | GAACTGGTCCCGATGTCC | 8124-8142 |
| GPgV F4 | To confirm the genome | This study | TGACTTGCGCATAACCCGCA | 7702-7721 |
| GPgV R4 | To confirm the genome | This study | AATCTCCGTTTCACGCTCTA | 9819-9838 |
| GPgV F5 | To confirm the genome | This study | TTACAGACTCTGGACGAGAT | 9491-9510 |
| GPgV R5 | To confirm the genome | This study | CACCTGGGCTAACCATACC | 11050-11068 |
| GPgV F6 | qRT-PCR; nested RT-PCR | This study | CGACAAGGGTGCCTGCTGA | 4620-4638 |
| GPgV R6 | qRT-PCR; nested RT-PCR | This study | GCCGTAGGTCGCATAGGT | 4796-4813 |
| Parvovirus-F1 | qPCR | 22 | TGCCGATGGAGTGGGTAAT | 3014-3032 |
| Parvovirus-R1 | qPCR | 22 | GTAGATGTGGTTGTTGTAGC | 3135-3134 |
| Parvovirus-F2 | PCR | This study | TATAGATAGCCTCCAACG | 2699-2716 |
| Parvovirus-R2 | PCR | This study | GGCATATACATCCGACGG | 3459-3476 |
| Circovirus-F | PCR | This study | CBATTAATAACCCTACCTTTGA | 116-137 |
| Circovirus-R | PCR | This study | GACCAATCAGAACGATGACC | 558-577 |
| Astrovirus-F1 | Nested RT-PCR | This study | ATGAATTRKATTBKYACAGCAG | 1766-1787 |
| Astrovirus-R1 | Nested RT-PCR | This study | KTCCATCHAYARTACACCA | 2024-2042 |
| Astrovirus-F2 | Nested RT-PCR | This study | GTGAAAACATCTGTGTTCGC | 1827-1846 |
| Astrovirus-R2 | Nested RT-PCR | This study | GTTTTGGAAGTTGGCATCCC | 1986-2005 |
| Coronavirus-F | RT-PCR | This study | ACTCARWTGAATTTGAAATAYGC | 13633-13655 |
| Coronavirus-R | RT-PCR | This study | TCACAYTTWGGATARTCCCA | 13864-13883 |
| Paramyxovirus-F | RT-PCR | This study | TCACTCCTCTTGGCGACTC | 4824-4842 |
| Paramyxovirus-R | RT-PCR | This study | CAAACTGCTGCATCTTCC | 5088-5105 |
| Avian Influenza Virus-F | RT-PCR | This study | TTCTAACCGAGGTCGAAAC | 33-51(M) |
| Avian Influenza Virus-R | RT-PCR | This study | AAGCGTCTACGCTGCAGTCC | 242-261(M) |
| Avian Influenza Virus-Uni12b | Reverse transcription | 23 | AGCAAAAGCAGG | — |
| GPgV-spreverse1 | Negative-strand RNA RT primer | This study | AAGCGGTATGCTGAGGG | 9884-9900 |
| GPgV-spreverse2 | Negative-strand RNA RT primer | This study | GTGCGTTGCCCCCTAAAA | 8852-8869 |
| GPgV-spreverse3 | Negative-strand RNA RT primer | This study | ACAACCAGTTCCGCTACC | 7941-7958 |
| GPgV-spreverse4 | Negative-strand RNA RT primer | This study | CTACACAGGGAAGTGGGC | 6937-6954 |
| GPgV-spreverse5 | Negative-strand RNA RT primer | This study | GGAAGACTGTCTCAGGAG | 5990-6007 |
| GPgV-spreverse6 | Negative-strand RNA RT primer | This study | GAGGAAGAGCGACTGCTC | 5009-5026 |
| GPgV-spreverse7 | Negative-strand RNA RT primer | This study | TGGGAAAAATGCTTTTGT | 4021-4038 |
| GPgV-spreverse8 | Negative-strand RNA RT primer | This study | AATGGAAAGATGTTGGCA | 3023-3040 |
| GPgV-spreverse9 | Negative-strand RNA RT primer | This study | AGTGGGTTCTGTTAGGAT | 2044-2061 |
| GPgV-spreverse10 | Negative-strand RNA RT primer | This study | TGCGAGTACCTGTCGCAA | 1065-1082 |
| GPgV-spreverse11 | Negative-strand RNA RT primer | This study | TCGGCACCATGGCGACCC | 561-578 |
| GPgV-spreverse12 | Negative-strand RNA RT primer | This study | TCCGCGGGTTGCCAATCC | 365-382 |
| GPgV-spreverse13 | Negative-strand RNA RT primer | This study | CGGCCCGAAAGGGCCGCG | 1-18 |
M, M gene; —, unfixed position.
Uni12 is a universal reverse transcription primer for influenza virus.
Characterization of the GPgV genome.
The GPgV-1 genome has a length of 11,479 nt, which is longer than the genome of any known pegivirus. It has relatively long 5′ and 3′ UTRs and a single ORF (Fig. 2A). The prediction of the RNA secondary structure for the first 586 nucleotides of the GPgV-1 genome revealed the presence of conserved IRES motifs (7): a GNRA motif (residues 511 to 514) is located in stem-loop X, and a polypyrimidine tract (PPT, residues 525 to 536) overlaps with stem-loop XII of the GPgV-1 5′ UTR (Fig. 1). Based on the location of these motifs, it was predicted that the AUG codon at position 569 is the most likely initiation codon and that the polyprotein would have the N-terminal sequence MATLVVFVLLALCPAG. The proposed position of the initiation codon is also consistent with previous observations that the N terminus of the pegivirus polyprotein contains leucine-rich motifs (6, 8–10). Thus, the 5′ UTR of GPgV-1 most likely has a length of 568 nt. The 3′ UTR has a length of 618 nt (Fig. 2A) and contains no polyU/UC element. The highest sequence identity (43.93%) for the 5′ UTR of GPgV-1 was found with its counterpart from pegiviruses infecting New World monkeys in the species Pegivirus A. For the 3′ UTR of GPgV-1, the closest match was the 3′ UTR of equine Pegivirus E (38.05% sequence identity).
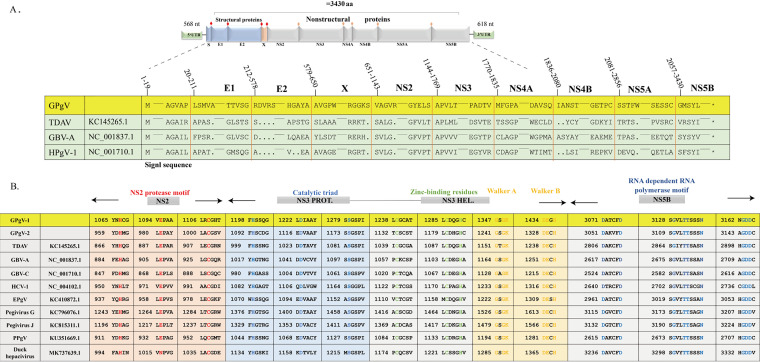
FIG 2.
Characterization of the GPgV genome and polyprotein. (A) GPgV-1 genome and polyprotein organization. Putative cleavage sites within the polyprotein are indicated by a red rhombus (structural proteins) or an orange rhombus (nonstructural proteins). Sequence alignment shows a comparison of the predicted cleavage sites for cellular signalase and viral proteases in the GPgV-1 polyprotein with other pegiviruses. (B) Locations and sequence comparisons of the conserved NS2 protease motif, NS3 protease catalytic triad, zinc-binding residues, Walker A, Walker B, and NS5B RNA-dependent RNA polymerase motifs.
The coding region of GPgV-1 also showed evidence for extensive RNA secondary structure, a feature previously associated with virus persistence in mammalian viruses (11, 12). The presence of RNA secondary structure was demonstrated through estimating minimum folding energy (MFE) differences between the native sequence and the mean of 49 sequence order randomized control sequences. These were generated by the algorithm NDR, which randomized nucleotide sequence order while retaining native dinucleotide frequencies (13). The GPgV-1 genome possessed a 10.7% mean MFE difference (MFED) over the coding region, similar to values obtained for pegiviruses infecting a range of mammalian species (2, 10).
The GPgV-1 genome encodes a polyprotein with a length of 3,430 amino acid residues. Sequence comparison with known pegiviruses indicates that this polyprotein encodes four structural proteins (a truncated protein S corresponding to Flavivirus core protein, E1, E2, and X) and six nonstructural (NS) proteins (NS2, NS3, NS4A, NS4B, NS5A, and NS5B) (Fig. 2A). A search for signalase cleavage sites (http://www.cbs.dtu.dk/services/SignalP/) revealed four sites located at amino acid residues 19, 211, 578, and 650. Sites for viral proteases were found in the expected positions and shared sequence similarity with those found in polyproteins of known pegiviruses (Fig. 2A). Similarly, many sequence motifs that are conserved in hepaciviruses and pegiviruses were also found in the GPgV-1 polyprotein, including the motif for NS2 cysteine proteases, the catalytic triad of serine proteases, zinc-binding residues characteristic of RNA helicases, and Walker A and Walker B motifs in NS3 (Fig. 2B). GPgV-1 NS5B consists of 574 amino acid resides and contains conserved motifs characteristic of RNA-dependent RNA polymerases (Fig. 2B).
Phylogenetic analyses of the GPgV sequence.
Genus Pegivirus consists of two distinct lineages, named groups 1 and 2 (2), which were also apparent in the analysis performed (Fig. 3A). Group 1 comprises HPgV-1 and viruses from nonhuman primates, pigs, and horses, while the HPgV-2 and pegiviruses from rodents fall into group 2. In addition, there are bat pegiviruses in both groups. To investigate the genetic relationship of GPgV with known pegiviruses, separate phylogenetic trees were constructed based on putative NS3 and NS5B regions (Fig. 3A). It was found that GPgV-1 and GPgV-2 form a unique branch (group 3) within the Pegivirus genus (Fig. 3A).
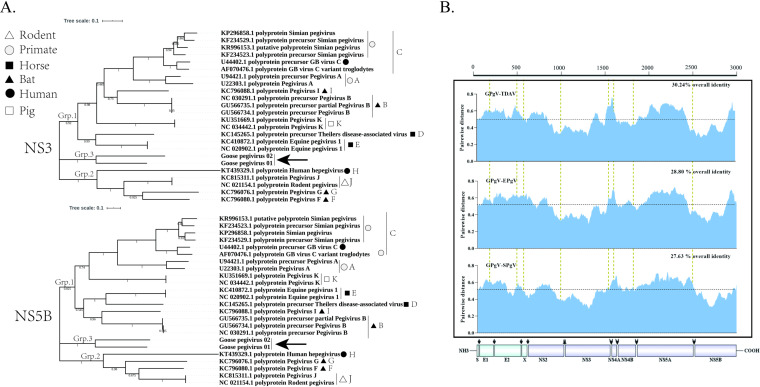
FIG 3.
Phylogenetic analysis of GPgV relative to other pegiviruses. Phylogenetic analysis was carried out using MEGA 7 software. (A) Phylogenetic trees of GPgV and representative strains from the Pegivirus genus built using the neighbor-joining method based on the sequences of NS3 (upper panel) and NS5B (lower panel) proteins. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (200 replicates) is shown next to the branches. The evolutionary distances were computed using the JTT matrix-based method and are presented in units of the number of amino acid substitutions per site. (B) Amino acid sequence divergence between polyproteins of GPgV, TDAV, EPgV, and SPgV. The sliding window size is 50 amino acid residues.
Next, the sequences of polyproteins encoded by GPgV-1, TDAV, equine pegivirus (EPgV), and simian pegivirus (SPgV) were aligned, and pairwise distances were computed. The obtained pairwise distance plots (Fig. 3B) showed that the regions with the lowest distances were mapped to the putative serine protease region of NS3 and the NS5B region (Fig. 3B), indicating that these proteins are the most conserved within the Pegivirus genus. The overall sequence identity between polyproteins of GPgV-1 and other pegiviruses was from 27.63% to 30.24% for group 1 pegiviruses and from 22.86% to 27.28% for group 2 pegiviruses; conservation was somewhat higher for nonstructural proteins than for structural proteins (Table 2). The highest sequence identities found for any individual protein were 51.27% with NS3 of EPgV and 52.81% with NS5B of bat pegivirus (Table 2).
TABLE 2.
UTR and polyproytein sequence identity between GPgV-1 and other pegiviruses
| Identity |
Pegivirus species |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Group I |
Group II |
||||||||||
| A | B | C | D | E | I | K | F | G | H | J | |
| % Nucleotide identity | |||||||||||
| GenBank accession no. | HGU22303 | GU566734 | AY196904 | KC145265 | KC410872 | KC796088 | KU351669 | KC796080 | KC796076 | KT439329 | NC_021154 |
| 5' UTR (568nt) | 43.93 (593nt) | 3.7 (46nt) | 38.85 (513nt) | 44.34 (617nt) | 37.74 (498nt) | 45.56 (566nt) | 39.26 (617nt) | 16.67 (216nt) | 3.35 (49nt) | 24.69 (329nt) | 25.26 (349nt) |
| 3' UTR (618nt) | 14.84 (195nt) | 18.87 (263nt) | 16.61 (267nt) | 18.97 (292nt) | 38.05 (664nt) | 8.23 (100nt) | 15.48 (224nt) | 17.74 (271nt) | 21.45 (308nt) | 2.75 (35nt) | 34.41 (475nt) |
| % Amino acid identity | |||||||||||
| GenBank accession no. | AAC55983 | ADK12629 | AAO42519 | AGH70217 | AGI04301 | AGK41018 | ANO81671 | AGK41010 | AGK41006 | ALE27082 | YP_007905734 |
| Polyprotein | 27.72 | 28.74 | 27.63 | 30.24 | 28.80 | 28.58 | 27.64 | 26.69 | 27.28 | 22.86 | 25.87 |
| Structural region | 19.84 | 18.08 | 25.61 | 18.77 | 17.40 | 19.16 | 17.80 | 15.09 | 13.09 | 14.43 | 12.44 |
| Nonstructural region | 29.73 | 30.68 | 28.38 | 31.66 | 31.68 | 30.01 | 29.49 | 28.75 | 30.07 | 24.12 | 28.35 |
| NS3 | 48.33 | 49.52 | 47.38 | 50.48 | 51.27 | 50.95 | 47.15 | 46.10 | 46.51 | 39.84 | 45.7 |
| NS5B | 44.64 | 52.30 | 46.02 | 50.42 | 47.91 | 52.81 | 48.19 | 46.02 | 47.92 | 36.68 | 44.98 |
GPgV infection in goslings results in reduced weight gain and high lymphotropism.
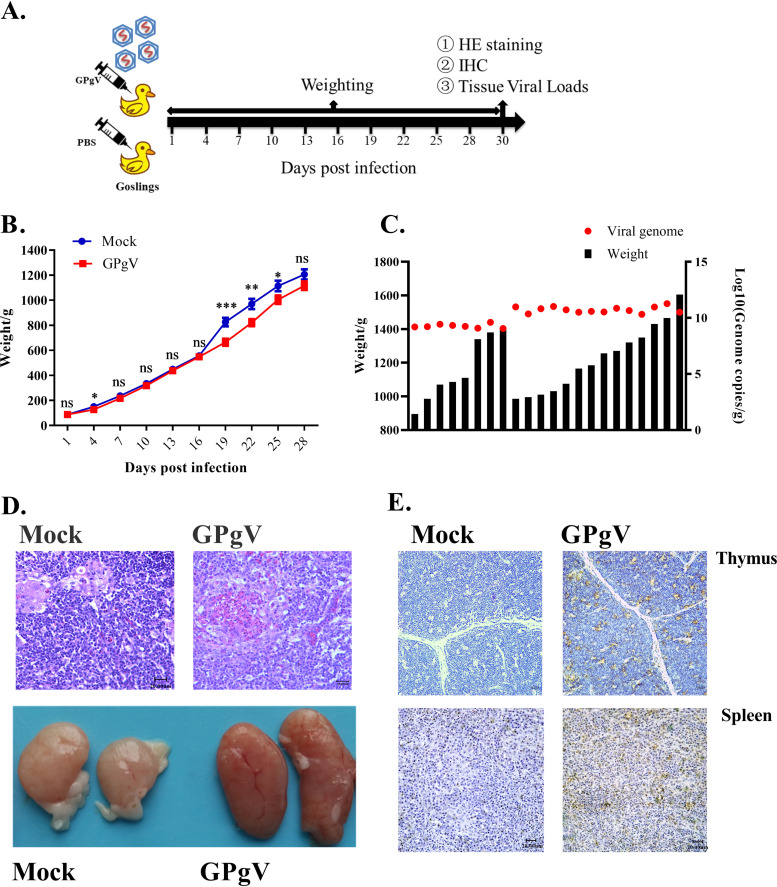
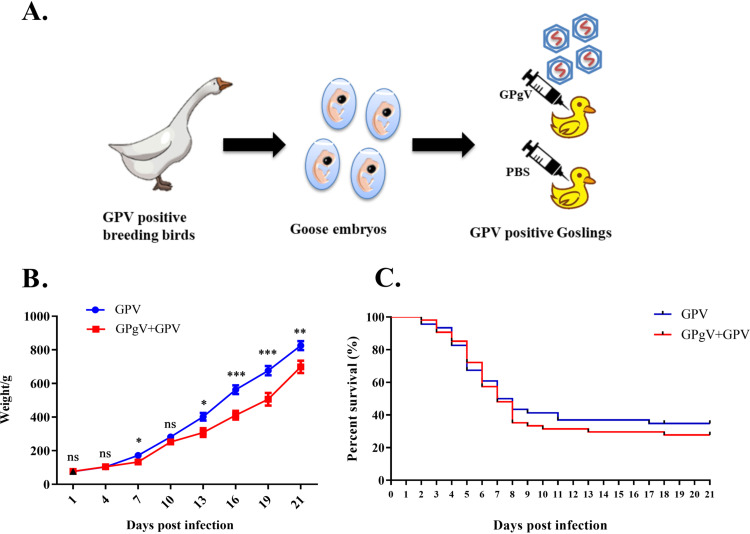
As no specific-pathogen-free (SPF) geese were available, the breeding birds were immunized with a live-attenuated GPV vaccine. Goslings from these parents were confirmed to be negative for known viruses and GPgV and used in this study. Twenty-four conventionally reared 3-day-old goslings were infected by intravenous inoculation of the allantoic fluid of GPgV-infected embryonated goose eggs. The control group of 20 goslings was mock infected. Infected/mock-infected goslings were kept in a waterfowl isolator for 30 days. All 20 goslings in the control group survived and tested negative for GPgV by nested RT-PCR at the end of the experiment (Fig. 4A). Over the course of the study, two infected goslings died, one at postinfection day (PID) 8 and another at PID 18. The gosling that died at PID 8 had undetectable GPgV RNA levels, while the gosling that died at PID 18 tested positive by quantitative reverse transcription-PCR (qRT-PCR). GPgV infection was found to significantly retard weight gain in the goslings at PID 19, 22, and 25; however, at PID 28 the effect was no longer significant (Fig. 4B). When the birds were sacrificed at PID 30, 6 out of 22 infected goslings showed moderate enlargement of the spleen, and 9 out of 22 infected goslings showed a hyperemic and moderately swollen thymus (Fig. 4D shows hematoxylin and eosin [HE] staining and the gross findings). The viral genome copy numbers in the spleens of infected goslings ranged from 1.15 × 109 to 1.87 × 1011 copies/g; no correlation between virus titer in the spleen and weight of the birds was observed (Fig. 4C). Interestingly, the gosling that died at PID 18 showed the highest GPgV genome copy number (5.22 × 1011 copies/g) in the spleen. Next, this gosling, as well as 5 goslings with the highest and 5 goslings with the lowest virus genome copy numbers in the spleen, were further analyzed for virus tissue distribution. Highest viral loads were found in the lymphoid tissues, such as the spleen and thymus (Table 3). GPgV infection was also visualized in the thymus and spleen of infected goslings by an immunohistochemistry (IHC) assay (Fig. 4E). However, a correlation between viral load and the weight gain decrease was not apparent.
FIG 4.
GPgV reduces weight gain and results in changes in the thymus of infected goslings. (A) Schematic representation of the study design. Three-day-old goslings from the experimental group (n = 24) were infected intravenously with 200 μl of allantoic fluid from GPgV-infected goose embryos (1.16 × 1010 RNA copies/ml), while goslings from the control group (n = 20) were mock infected with 200 μl PBS. Every gosling was weighed at PID 1, 4, 7, 10, 13, 16, 19, 22, 25, and 28. At 30 PID, surviving birds were sacrificed and spleen, thymus, bursa of Fabricius, duodenum, rectum, pancreas, and liver were sampled for HE staining, ICH, and tissue viral load detection. (B) Goslings were weighed at the indicated days postinfection. Mean values are shown, and error bars indicate the standard deviation: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant (Student’s t test). (C) The viral RNA copy numbers in the spleen (red dots) and the body weights (black column) of the infected goslings at PID 30. (D) Upper: representative HE staining of the thymus in the control group (left) and experimental group (right) at PID 30. Bars = 20 μm. Lower: the gross findings of the thymus from mock-infected (left) and GPgV-infected geese (right) at PID 30. (E) The detection of GPgV in the thymus and spleen of infected goslings at PID 30 using an anti-GPgV E2 monoclonal antibody. Bars = 20 μm.
TABLE 3.
Tissue viral loads of GPgV in experimental inoculation studya
| PID | Viral loads in spleen | Tissue viral load (log10 copies/g) |
||||||
|---|---|---|---|---|---|---|---|---|
| Spleen | Thymus | Bursa of Fabricius | Rectum | Duodenum | Pancreas | Liver | ||
| 18 (dead gosling) | 11.72 | 10.85 | 11.48 | 11.13 | 11.25 | 10.12 | 10.42 | |
| 30 | Top 5 values | 11.27 | 11.28 | 11.22 | 10.77 | 10.28 | 10.23 | 9.95 |
| 11.02 | 10.43 | 10.41 | 10.92 | 10.41 | 9.12 | 8.93 | ||
| 10.98 | 10.48 | 10.59 | 10.38 | 10.17 | 9.51 | 9.53 | ||
| 10.97 | 10.95 | 10.63 | 9.79 | 10.21 | 9.05 | 8.98 | ||
| 10.86 | ND | 10.67 | 10.12 | 10.26 | 8.91 | 9.38 | ||
| Bottom 5 values | 9.24 | ND | ND | 9.12 | ND | ND | ND | |
| 9.22 | 10.52 | 9.36 | 9.00 | ND | 9.21 | ND | ||
| 9.19 | 9.63 | 9.44 | 8.92 | 9.02 | ND | ND | ||
| 9.08 | 10.17 | 10.76 | 8.78 | 9.29 | ND | ND | ||
| 9.06 | 10.41 | 9.65 | 9.93 | 9.49 | ND | ND | ||
PID, postinfection day; ND, negative detection.
GPgV coinfection increases the weight gain reduction and mortality of goslings infected with GPV.
Coinfections are frequent in both humans and animals. As pegiviruses have been reported to modify the outcomes of infections caused by distantly related and unrelated viruses (14–16), the dynamics of GPgV infection in the case of coinfection with GPV were analyzed. In these experiments, 54 goslings confirmed to be GPV positive by PCR were infected with GPgV as described above, and 46 GPV-positive goslings were mock infected (Fig. 5A). The birds were kept in a rearing isolator for 3 weeks and observed as described above. Reduced feed intake and profound growth retardation were observed for GPgV+GPV coinfected birds; from PID 13, the body weights of surviving coinfected goslings were significantly lower than the body weights of surviving control goslings (Fig. 5B). As expected, GPV infection alone resulted in the death of most goslings within 18 days; the mortality rate was 65.22% (30 out of 46). The mortality of GPgV+GPV coinfection was slightly higher (72.22%) (39 out of 54) but the difference was not statistically significant (Fig. 5C). In the coinfected group, 64.81% (35/54) of goslings were GPgV positive, including all birds that survived until PID 21. Very high levels of GPgV RNA (up to 4.16 × 1011 copies/g) were detected in the spleens of these birds. The seemingly modest coinfection rate was probably because 15 (27.78%) of the coinfected goslings died before PID 5, i.e., before GPgV RNA levels became detectable. Samples from 8 goslings that died at PID 5 to 8 and were confirmed to be double positive (GPgV+GPV) were compared with samples from the same number of birds from the control group (GPV alone) that died at the same time. Similar to the birds infected with only GPgV, high viral loads were detected in the thymus (Table 4). GPV genome copy numbers varied from 2.29 × 109 (pancreas) to 2.22 × 1012 (spleen) copies/g in coinfected goslings, while levels of 4.27 × 108 (pancreas) to 4.16 × 1012 (spleen) copies/g were observed in the control group; there was no clear difference between the two experimental groups (Table 5).
FIG 5.
GPgV coinfection reduces weight gain and survival of GPV-carrying goslings. (A) Schematic representation of the study design. Two-day-old GPV-positive goslings (n = 54) were intravenously infected with 200 μl of allantoic fluid containing 1.16 × 1010 GPgV copies/ml, while the control group (n = 46) was mock infected with 200 μl of PBS. (B) Three randomly selected live goslings from both groups were weighed at the indicated days postinfection. Mean values are shown, and the error bars indicate the standard deviation: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant (Student’s t test). (C) Kaplan-Meier survival plot of GPV-infected and GPgV+GPV-coinfected goslings; birds were monitored up to 21 days postinfection.
TABLE 4.
Tissue viral loads of GPgV in the dead goslings of experimental coinfection studya
| PID | Tissue viral load (log10 copies/g) |
||||||
|---|---|---|---|---|---|---|---|
| Spleen | Thymus | Bursa of Fabricius | Rectum | Duodenum | Pancreas | Liver | |
| 6 | 8.74 | 9.25 | 9.67 | 9.28 | ND | ND | ND |
| 7 | 9.42 | 9.61 | 8.71 | ND | 9.22 | ND | ND |
| 9.36 | 9.28 | 8.92 | ND | 8.71 | ND | ND | |
| 9.19 | 9.65 | ND | ND | 9.35 | 8.95 | ND | |
| 8.94 | 9.10 | 8.87 | ND | ND | 8.90 | 8.95 | |
| 8 | 9.24 | 10.74 | ND | 9.90 | 9.63 | ND | ND |
| 9.31 | 9.41 | 9.06 | 9.27 | 8.77 | 8.97 | ND | |
| 9.02 | 9.15 | ND | 9.34 | 9.91 | ND | ND | |
PID, postinfection day; ND, negative detection.
TABLE 5.
Tissue viral loads of GPV in the dead goslings of experimental coinfection studya
| Group | PID | Tissue viral load (log10 copies/g) |
||||||
|---|---|---|---|---|---|---|---|---|
| Spleen | Bursa of Fabricius | Thymus | Rectum | Duodenum | Pancreas | Liver | ||
|
Coinfection GPgV+/GPV+ |
6 | 11.55 | 11.36 | 12.01 | 11.64 | 10.70 | 9.55 | 11.32 |
| 7 | 11.77 | 10.48 | 11.86 | 11.72 | 11.53 | 10.35 | 11.18 | |
| 11.43 | 10.09 | 10.42 | 11.35 | 11.32 | 9.56 | 12.16 | ||
| 12.35 | 10.48 | 11.34 | 11.16 | 10.00 | 10.18 | 11.87 | ||
| 11.84 | 11.73 | 11.34 | 11.24 | 11.74 | 10.10 | 11.47 | ||
| 8 | 12.21 | 11.54 | 11.39 | 11.34 | 11.43 | 9.93 | 12.35 | |
| 12.19 | 10.25 | 11.29 | 11.47 | 11.41 | 9.36 | 11.52 | ||
| 12.12 | 11.36 | 11.51 | 12.13 | 11.60 | 10.34 | 10.51 | ||
| Single infection GPV+ | 5 | 12.58 | 11.04 | 11.96 | 12.01 | 11.96 | 10.82 | 11.86 |
| 6 | 11.58 | 10.60 | 11.62 | 11.27 | 11.48 | 10.61 | 11.06 | |
| 7 | 12.24 | 11.49 | 11.49 | 12.52 | 12.30 | 11.06 | 11.28 | |
| 12.62 | 11.14 | 12.41 | 11.72 | 11.80 | 10.72 | 12.25 | ||
| 11.21 | 11.04 | 10.91 | 11.80 | 11.77 | 10.94 | 10.95 | ||
| 8 | 11.84 | 11.06 | 11.03 | 11.67 | 11.82 | 10.60 | 11.02 | |
| 12.08 | 10.80 | 10.81 | 11.54 | 11.70 | 10.58 | 11.69 | ||
| 11.77 | 10.53 | 11.29 | 12.52 | 11.65 | 8.63 | 11.43 | ||
PID, postinfection day.
Passaging of GPgV in infected embryonated goose eggs does not affect virulence but slightly increases virus replication.
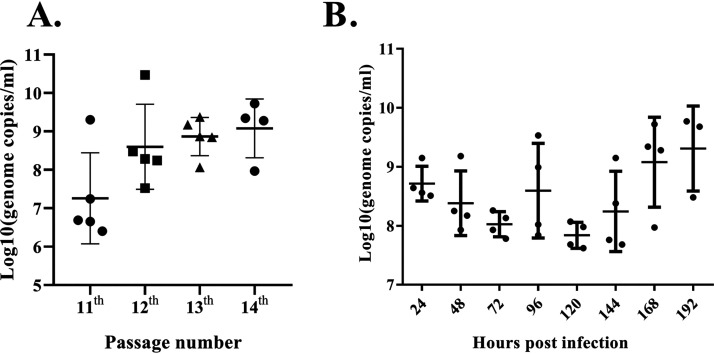
The virus stock used for identification of the GPgV (i.e., stock obtained after 10 serial passages in 9-day-old embryonated goose eggs) was passaged four more times. Albeit passages were generally performed using allantoic fluid from dead eggs, no increase of the mortality of infected embryos was observed: during both initial (passages 1 to 10) and subsequent (passages 11 to 14) passaging mortality varied from 0 to 66.67% and the death of infected embryos mainly occurred from 59 to 174 h postinoculation. Thus, the GPgV virulence (the mortality rate of GPgV-infected embryonated goose eggs) did not increase with increasing passage numbers. GPgV genome copy numbers in the allantoic fluid varied from 1.83 × 106 to 2.18 × 109 copies/ml in the dead eggs and from 1.83 × 106 to 2.95 × 1010 copies/ml in the eggs that survived. In contrast to the virulence, the proliferation of GPgV in the 11th, 12th, 13th, and 14th passages increased (Fig. 6A). Nevertheless, as the GPgV genome copy numbers in the allantoic fluid of individual embryonated eggs showed large variations, the observed increase was not statistically significant. Importantly, similar to what was observed in GPgV-infected goslings in vivo, there was no strong correlation between the extent of viral replication and death caused by infection.
FIG 6.
Growth characteristics of GPgV in 9-day-old embryonated goose eggs. (A) GPgV stock (10th generation) was passaged in 9-day-old embryonated goose eggs four consecutive times; for each passage, allantoic fluid was harvested at 168 h p.i. The virus genome copy number in the 11th, 12th, 13th, and 14th generation stocks was determined by qRT-PCR. Each symbol represents an individual egg (n = 5, 5, 5, 4 for each generation, respectively); the line depicts the mean of each group, and the error bars indicate the standard deviation. (B) GPgV from the 14th passage (200 μl, 4.55 × 109 RNA copies/ml) was inoculated into embryonated eggs (n = 4 for each time point). At the indicated time points, virus genome copy numbers in the allantoic fluid were determined by qRT-PCR; data are presented as described for panel A.
The time course of GPgV proliferation in embryonated eggs was also analyzed. Eggs were infected with GPgV from the 14th passage, and genome copy numbers were determined every 24 h from 24 h to 192 h postinfection (p.i.). Again, due to the large variation in virus genome copy numbers between individual infected eggs, a classical viral growth curve was not observed. However, the trend was that at first the average viral load slightly declined, and then, from 120 h p.i., the viral load gradually increased with incubation time and reached the highest values by the end of the experiment (Fig. 6B).
GPgV proliferates well in primary GEFs but not in other cell types.
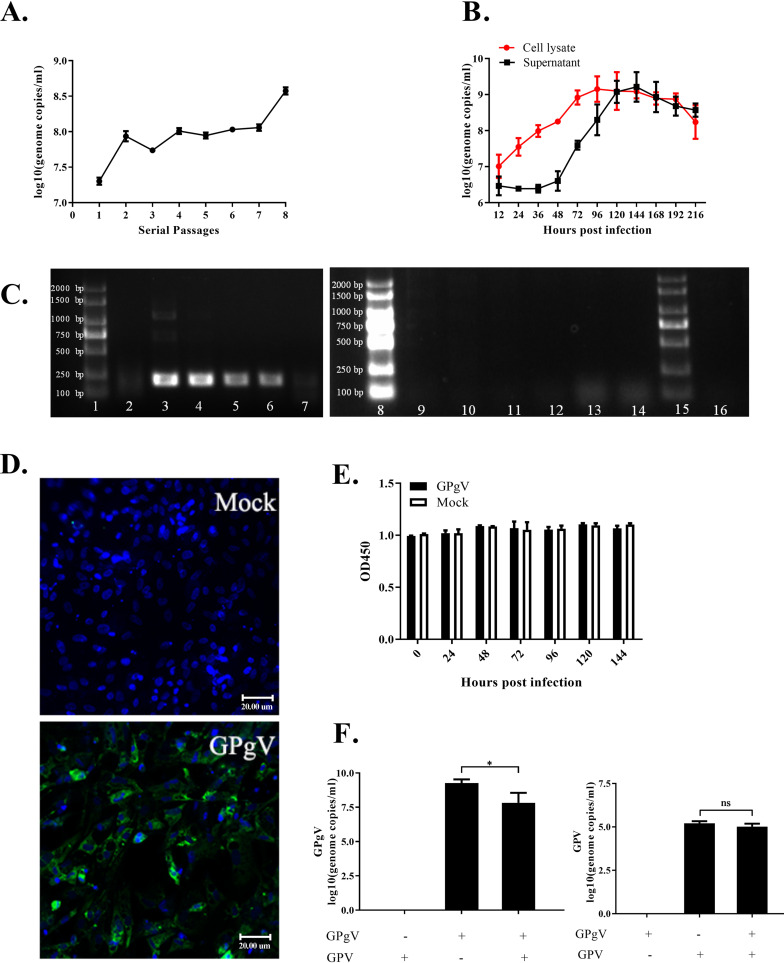
Primary goose embryonic fibroblasts (GEFs) were used to establish a cell culture model for GPgV infection. GEFs were infected with GPgV using allantoic fluid from GPgV-infected goose embryos that contained 2.99 × 108 GPgV genome copies/ml. The virus was passaged 8 times in GEFs; in no passage were any cytopathic effects (CPE) observed. At the same time, GPgV genome copies in the cell culture supernatants gradually increased from 2.02 × 107 copies/ml (passage 1) to 3.78 × 108 copies/ml (passage 8), indicating some cell culture adaptation of virus (Fig. 7A).
FIG 7.
Growth characteristics of GPgV in GEFs. (A) GPgV derived from allantoic fluid was passaged 8 times in GEFs. The virus genome copy number in the cell culture supernatant was measured for every passage. (B) The GEFs were infected with cell-culture-adapted GPgV (7th passage, 1.15 × 108 RNA copies/ml) and, at the indicated times, culture supernatants and cells lysates, which were obtained using three freeze-thaw cycles, were collected. The virus genome copy number was determined by qRT-PCR. The cell lysate indicates amount of viral RNAs in 1 ml of lysate. (C) GPgV negative-strand RNA assay. GPgV-spreverse primers were used for a reverse transcription reaction of GPgV negative-strand RNA assay. H2O (no primer) used for a reverse transcription reaction functioned as the blank control. Nested RT-PCR using in vitro-transcribed RNA for a template functioned as the negative control. Lanes: 1, marker; 2, GEF-1; 3, GEF-4; 4, GEF-8; 5, thymus; 6, spleen; 7, small intestine. 8, marker; 9, GEF-1; 10, GEF-4; 11, GEF-8; 12, thymus; 13, spleen; 14, small intestine. 15, marker; 16, positive-sense RNA. (D) Detection of the GPgV E2 protein in GEFs at 96 h p.i. Control cells (Mock) were mock infected with PBS. The samples were treated with a mouse anti-GPgV E2 monoclonal antibody as the primary antibody and FITC goat anti-mouse IgG as the secondary antibody (green); nuclei were counterstained with DAPI (blue). Fluorescence was detected using a Nikon 80i microscope. Scale bars indicate 20 μm. (E) The viability of GPgV-infected and mock-infected GEFs was tested by a CCK8 assay. (F) GEFs grown in a 6-well plate were infected with GPgV (7th passage, 1.15 × 108 RNA copies/ml) and/or GPV (6.46 × 106 DNA copies/ml). The numbers of GPgV genome copies (left) and GPV DNA copies (right) were determined at 168 h p.i. All the experiment was repeated three times. Mean values are shown, error bars indicate the standard deviation: *, P < 0.05; ns, not significant (Student’s t test).
The 8th passage GPgV stock was used to obtain a growth curve of GPgV in cell culture. It was observed that in the lysates of infected cells, the viral genome copy number gradually increased with incubation time and reached a plateau at 72 h p.i. In the cell culture supernatant, no increase in virus genome copy number could be detected up to 48 h p.i. From this point onward, the viral genome copy number increased rapidly and reached a plateau at 120 h p.i. indicating the release of progeny viruses (Fig. 7B). Negative-strand PCR assays were used to confirm virus replication in cell culture (Fig. 7C, lanes 2 to 4), and also in the spleen, thymus, and small intestine of the infected goslings at PID 30 (Fig. 7C, lanes 5 to 7). All results were strongly positive except for weak positives in the first generation of GPgV-infected GEFs and in the small intestine (Fig. 7C, lanes 2 and 7). Reactions with no reverse primers were used as controls (Fig. 7C, lanes 9 to 14), and the positive-strand RNA used as a negative control confirmed the assay was specific (Fig. 7C, lane 16). The expression of the GPgV E2 protein in infected GEFs was clearly detected by an indirect immunofluorescence assay (Fig. 7D). Analysis of the viability of GPgV-infected GEFs using a CCK8 cytotoxicity assay revealed no decrease in cell viability compared to that of noninfected cells (Fig. 7E); this finding is consistent with the lack of visually detectable CPE.
Similar to in vivo experiments, we also performed an analysis of GPgV and GPV coinfection in GEFs. There was no significant difference in GPV genome copy numbers in the presence of absence of GPgV coinfection (Fig. 7F, right panel), which is similar to the observations made in vivo. In addition, this analysis revealed that at 168 h p.i., the genome copy number of GPgV was significantly lower in GPgV+GPV coinfected cells than in cells infected with GPgV alone (Fig. 7F, left panel). Thus, under in vitro cell culture conditions, GPV inhibits GPgV replication but not vice versa.
In order to determine whether GPgV can also infect cells from different hosts, we used duck embryonic fibroblasts (DEFs), baby hamster kidney cells (BHK-21), and bat lung cells (Tb1Lu) and performed the passaging experiments as described for GEFs. Similar to GEFs, no CPE was observed in any cell line during the 96 h observation period. However, in contrast to GEFs (Fig. 7A), we were unable to detect any GPgV RNA in DEFs, BHK-21 cells, and Tb1Lu cells from the 2nd passage and onward (Table 6), clearly indicating that these cells do not support GPgV infection. Similar results were also obtained for primary goose peripheral blood lymphocytes (GPBLs) and spleen lymphocytes (GSLs). Although these cells are of goose origin, no GPgV adaptation was observed, and GPgV RNA levels were below the level of detection by the 2nd and 3rd passages in GPBLs and GSLs, respectively (Table 6). Thus, GPgV has high host and cell-type specificity, and the only cells where the virus was able to propagate were GEFs (Fig. 7B).
TABLE 6.
Detection of GPgV RNA in different cell types
| Passage | Viral genome (log10 copies/ml)a |
||||
|---|---|---|---|---|---|
| DEF | BHK-21 | Tb1Lu | GPBL | GSL | |
| F1 | 7.82 | 7.42 | 6.96 | 6.39 | 7.81 |
| F2 | ND | ND | ND | ND | 6.84 |
| F3 | ND | ND | ND | - | ND |
| F4 | ND | ND | ND | - | - |
| F5 | ND | ND | ND | - | - |
ND, negative detection; -, no passage.
DISCUSSION
The evolutionary history of the Flaviviridae family is complex. Our understanding of the diversity of these viruses has significantly changed as new members of this family are discovered and characterized. Very recently, a duck hepacivirus-like virus with a pegivirus A-like IRES element in its 5′ UTR was reported (17). Here, a novel virus identified as a putative causative agent of disease in goslings and named GPgV, was, based on its genome structure, sequence identity, and phylogeny, classified as a new species within the genus Pegivirus. As the first pegivirus discovered in birds, GPgV also forms a unique branch within the pegiviruses that is distinct from previously described group 1 and 2 viruses. Discovery of GPgV serves as a proof that the host range of pegiviruses is not limited to mammals. Birds are the modern descendants of the dinosaurs, which fills the evolutionary gap between mammals and other vertebrates. Therefore, it is possible that the pegivirus host range extends further into different groups of vertebrates.
All currently known pegiviruses share an ability to persist in their natural hosts and are characterized by the lack of any identified pathogenicity (1, 2). The only possible exception is TDAV (a member of the Pegivirus E species). However, TDAV replication itself is not cytocidal, and clinical hepatitis, as observed upon experimental inoculation of horses with TDAV-positive plasma, is more likely to result from secondary immune or inflammatory mechanisms than directly from TDAV infection (3). In this context, it is interesting that GPgV was discovered as a result of searching for causative agents of goose disease. Similar to the case of TDAV, the role of GPgV in goose disease remains incompletely understood. On the one hand, no other potential pathogens were discovered in animals with the disease using different PCR-based methods and an unbiased NGS-based approach. We were also able to demonstrate that GPgV does, at least to some extent, correspond to Koch’s postulates: the use of cultured GPgV as an inoculum resulted in growth retardation and the virus showed high lymphotropism in the infected goslings. GPgV replication was confirmed in many tissues, with the highest viral loads found in immune tissues, such as the spleen and thymus. On the other hand, the clinical symptoms described in bird farms were not observed, and only one infected bird, which had a high GPgV load, died. Furthermore, a clear correlation between viral load, lymphotropism, and growth retardation was not apparent. Therefore, based on current data, it is difficult to state that GPgV is the causative agent of the original gosling disease. It is possible that another infectious agent was lost either during infectious material preparation or during passaging in embryonated goose eggs prior to the generation of libraries for NGS. Furthermore, the obtained NGS data cannot be used to detect the presence of an infectious agent that is highly divergent in sequence from any known virus.
The route of infection may also alter/prevent disease development/pathology. In an attempt to find the best route for experimental infection, we tested intramuscular and intravenous injection, as well as oral administration, of the allantoic fluid of GPgV-infected embryonated goose eggs. It was observed that GPgV could infect goslings via every infection route and replicated well in all cases. We detected a relatively high viral load in both the small and large intestine, but no GPgV RNA was detected in the limited feces samples of infected goslings. These data do not exclude the possibility of horizontal transmission of GPgV via contact with virus-contaminated feed and water. However, there are currently no data on horizontal or vertical transmission of the disease via infected breeders on farms. The question of whether fetal or neonatal hosts are sensitive to GPgV infection/disease due to their incompletely developed immune systems remains unanswered. Furthermore, the impact of additional factors or exposures that influence the course of disease associated with GPgV infections remains unknown. These and many other questions need to be addressed in further studies.
Outbreaks of gosling deaths are always associated with intestinal obstruction and intestinal necrosis. These symptoms were not observed upon GPgV infection; however, they are the classical clinical manifestations of GPV infection. In our study, though, GPgV coinfection did not boost GPV replication. This is similar to observations made with SPgV, which had no effect on simian immunodeficiency virus load in the acute phase of infection (4). Nevertheless, GPV+GPgV-coinfected goslings clearly showed elevated symptoms and had somewhat higher mortality than goslings infected with GPV alone. This finding contrasts with the lack of any identified pathogenicity of previously reported pegiviruses. The mechanism(s), possibly including GPgV infection showing lymphotropism, by which GPgV promotes GPV clinical manifestation, remain uncharacterized.
The thymus and spleen are the organs that bear the highest GPgV loads in infected goslings. The association of lymphotropism with GPgV infection is also clear. Therefore, it would be logical to expect that the detected lymphotropism may be caused by infection of lymphocytes. Nevertheless, GPgV failed to replicate in cultivated goose lymphocytes. This does not, however, exclude the possibility that the virus may replicate in T lymphocytes abundant in the thymus and spleen in vivo. Currently, we have no data on the interaction between GPgV and goose T lymphocytes; thus, specific studies will be needed to reveal the mechanism by which pegivirus affects host cellular immunity.
Surprisingly, GPgV could replicate well in embryonated goose eggs and adapt, at least to some extent, to growth in eggs. However, GPgV clearly did not proliferate identically in every embryonated egg (Fig. 6) and adaptation was also not associated with increased pathogenicity. In fact, the GPgV genome copy numbers in the allantoic fluid of the dead eggs were somewhat lower than those in the allantoic fluid of the viable eggs. Similarly, no clear correlation with infection time or the severity of clinical manifestations could be observed.
No cell culture model for a pegivirus has been previously reported. In this regard, it is important that GPgV replicates in primary GEFs. The virus could also be adapted to cell culture, which was not associated with the development of detectable CPE. An in vitro growth curve of GPgV has a standard shape (Fig. 7B) and revealed that the virus levels in the cell culture supernatant reached a plateau at 120 h p.i. and remained high after that time. This observation is consistent with persistent infection of pegiviruses in nature. Thus, GPgV is the first cell culture model for pegivirus, opening new possibilities for studies of pegivirus molecular biology.
Interestingly, GPgV failed to propagate in DEFs, although ducks are genetically close to geese. Similarly, our attempts to adapt GPgV for growth in 9-day-old embryonated duck eggs failed. Although GPgV is evolutionarily related to bat pegivirus and the bat has been proposed as a major natural reservoir for pegiviruses (18), no replication was observed in bat (Tb1Lu) cells. Taken together, these findings indicate high host-species specificity of GPgV. Furthermore, contrasting to the high in vivo titers observed in immune tissues, GPgV was unable to efficiently propagate in two types of goose lymphocytes, GPBLs and GSLs. This may be because the isolated lymphocytes have no self-multiplication ability and cannot be maintained in culture as long as primary fibroblasts. Collectively, our findings show that GPgV proliferates in cell culture in a species- and cell-type specific manner. Therefore, the cell culture model of GPgV infection can be used for studies of host determinants of GPgV infection and pegivirus host cell interactions in general.
MATERIALS AND METHODS
Sample collection and animal care and use.
Organ samples, including the duodenum, rectum, liver, kidney, spleen, and brain, were collected from dead geese in Sichuan Province and Chongqing municipality in China. In the gosling inoculation study, organ samples, including the spleen, thymus, bursa of Fabricius, duodenum, rectum, pancreas, and liver were collected from dead goslings or sacrificed goslings.
The animal studies were approved by the Institutional Animal Care and Use Committee of Sichuan Agricultural University and followed National Institutes of Health guidelines for the performance of animal experiments.
Nucleic acid extraction from tissues and cultured cells.
Total RNA was isolated using the RNAiso Plus reagent (TaKaRa, China) as follows. One milliliter of RNAiso Plus reagent was added per 10 mg organ sample, followed by homogenization. The homogenized sample was centrifuged at 12,000 × g for 5 min at 4°C. An aliquot of 100 μl of 1-bromo-2 chloropropane (BCP, Sigma-Aldrich, USA) was added to the obtained supernatant. The sample was shaken vigorously, incubated for 5 min at room temperature, and centrifuged at 12,000 × g for 10 min at 4°C. Isopropanol (500 μl) was added to the supernatant and the sample was incubated at room temperature for 10 min. After centrifugation at 12,000 × g for 10 min, the obtained pellet was washed with 1 ml of 75% ethanol, dried, and dissolved in 40 μl of H2O. To isolate nucleic acids from allantoic fluid, suspended cells, and homogenized organ samples, the TIANamp Virus DNA/RNA kit (Tiangen Biotech, China) was used according to the manufacturer’s instructions. Briefly, 200 μl of sample was treated with the provided reagents, and the obtained DNA/RNA was dissolved in 20 μl of H2O.
Virus detection.
The PCR assay for goose DNA viruses, including goose parvovirus and goose circovirus, contained 0.2 μl of DNA template, 0.4 μl of 10 μM mixed PCR primers (Table 1), 4.4 μl of H2O, and 5 μl of 2× Taq Master Mix (Vazyme, China). The qPCR assays for goose parvovirus contained 0.4 μl of DNA template, 0.6 μl of 10 μM mixed qPCR primers (Table 1), 4 μl H2O, and 5 μl of 2× Taq SYBRGreen qPCR Premix (Innovagene, China). qPCR assays were performed on a CFX96 Bio-Rad system (CFX96 Touch, Bio-Rad, USA).
One microgram of RNA was used as the template in 20-μl reverse transcription reaction mixtures containing RT-primer Mix and PrimeScrip RT reagent (TaKaRa, Dalian, China). For detection of avian influenza virus, the RT-primer Mix was replaced by avian influenza virus-Uni12 mix (Table 1). The RT-PCR for the detection of goose RNA viruses, including GPgV, goose astrovirus, goose coronaviruses, goose paramyxovirus, and avian influenza virus, contained 0.5 μl of cDNA template, 0.4 μl of 10 μM mixed PCR primers (Table 1), 4.1 μl of H2O, and 5 μl of 2× Taq Master Mix. In nested RT-PCR for GPgV and goose astrovirus, the second reaction contained 0.1 μl of the first PCR as a template (GPgV NF2-R2 for the first PCR and GPgV F6-R6 for the second PCR) (Table 1).
The SYBR green qRT-PCR assay was used to determine the GPgV genome copy numbers. Reaction mixtures contained 0.4 μl of cDNA template, 1 μl of 10 μM qPCR primer mix (Table 1), 3.6 μl of H2O, and 5 μl of PowerUp SYBR green Master mix (Life Technologies, USA); water was used as a blank control and materials from an uninfected probe as a negative control. Assays were performed using a CFX96 Bio-Rad instrument and the following parameters: 50°C for 2 min, 95°C for 2 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min, followed by dissociation analysis. In the data interpretation, the Cq value of >37 was regarded as a negative result; this value was determined with reference to the mean Cq value of the control group.
Library preparation and NGS.
Intestinal and liver tissues of diseased geese collected from Southwest China were homogenized in sterile phosphate-buffered saline (PBS) (pH 7.2). The obtained 20% suspension (wt/vol) was centrifuged at 7,000 × g at 4°C for 10 min, filtered through a 0.22-μm filter, and used to inoculate 9-day-old embryonated goose eggs (0.3 ml/egg) via the allantoic sac. Inoculated eggs were incubated at 37°C and observed daily. The allantoic fluid of embryos that died after 24 h or survived until 178 h after inoculation was collected. The putative pathogen was serially passaged 10 times, after which the allantoic fluid was collected and clarified by centrifugation at 7,000 × g for 20 min. The obtained supernatant (100 ml) was ultracentrifuged at 130,000 × g for 2 h at 4°C, and the obtained pellet was resuspended in 1 ml of PBS. DNA and RNA were extracted from the pellet and supernatants using a QIAamp MinElute virus vacuum kit (Qiagen, Germany). NGS was performed by BGI (Shenzhen, China). To obtain the sequence of the possible RNA virus genome, the reads from the RNA sample were compared to known sequences. Sequences not belonging to a novel pegivirus were removed, and the cleaned data were assembled using the SOAPdenovo (v1.05) short-sequence assembly software (19, 20). After multiple adjustments, the sample main parameter K was set to 59 and the reads were compared to the contig obtained by the assembly.
Genome-scale RNA structure prediction.
The GPgV-1 genome sequence was analyzed for evidence of genome-scale RNA structure (GORS) by comparing folding energies of consecutive fragments of nucleotide sequence with random sequence-order controls using the program Folding Energy Scan in the SSE package (13). Results were expressed as MFE differences (MFEDs), i.e., the percentage difference between the MFE of successive 240-base segments of the GPgV-1 coding sequence incrementing by 9 bases from that of the mean value of 49 sequence order-randomized controls (1,121 sequence fragments analyzed).
Immunofluorescence assays.
The mouse anti-GPgV E2 monoclonal antibody was prepared by Abmart Biomedical Co., Ltd. (China). Specificity of binding was verified using indirect enzyme-linked immunosorbent assay (ELISA) and Western blotting assays. GEFs obtained from 10-day-old goose embryos and maintained in minimal essential medium (MEM) with 10% fetal bovine serum (FBS) were seeded on coverslips placed in 12-well cell culture plates. At 70 to 90% confluence, cells were infected with GPgV (7th passage, 1.15 × 108 RNA copies/ml). At 72 h p.i., the cells were fixed in 4% paraformaldehyde for 1 h and permeabilized with 0.25% Triton X-100 for 1 h at 4°C. After three washes with PBS, the cells were blocked with 5% bovine serum albumin (BSA) in PBS at 37°C for 1 h, incubated overnight with anti-GPgV E2 antibody at 4°C, washed, and incubated with fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG antibody (Life Technologies, USA) at 37°C for 1 h. Nuclei were counterstained with 4’, 6-diamidino-2-phenylindole (DAPI, Coolaber, China). Fluorescence was examined using an 80i microscope (Nikon, Japan).
HE staining and immunohistochemistry.
The goose thymus tissues were fixed in 4% paraformaldehyde, dehydrated, embedded in paraffin, and sectioned into 4-μm-thick sections. The slides were stained with hematoxylin and eosin (HE) using standard procedures.
For immunohistochemistry, paraffin-embedded tissues were deparaffinized in xylene and rehydrated in graded alcohol. The obtained slides were boiled in 0.01 M citrate buffer for 20 min, treated with 3% H2O2 for 15 min at room temperature, and incubated in blocking solution containing 5% BSA, followed by overnight incubation at 4°C with mouse anti-GPgV E2 monoclonal antibody. The slides were incubated with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (Earthox, USA) for 1 h at 37°C, colored with diaminobenzidine solution (Solarbio, Beijing, China) for 4 min, and counterstained with hematoxylin at room temperature. Then, the slides were treated with graded alcohol and xylene and observed under an 80i microscope (Nikon, Japan).
Inoculation of embryonated eggs.
Virus stock obtained as described above was used to obtain the 11th, 12th, and 13th generations of the virus. Five eggs was used for each passage and only the allantoic fluid from the egg with the highest virus load, determined using qRT-PCR, was used for the next inoculation step. To obtain the allantoic fluid of the 14th generation, 32 eggs were inoculated and virus was collected at 24, 48, 72, 96, 120, 144, 168, and 192 h p.i. (4 eggs per each time point); virus loads were determined using qRT-PCR.
Cell culture experiments.
GEFs were infected using allantoic fluid from the 12th passage of the virus from embryonated eggs containing 2.99 × 108 copies of viral RNA/ml. The cells were incubated for 1 h, Dulbecco’s modified Eagle’s medium (DMEM, Life Technologies, China) containing 2% FBS was added. The cells were cultured for 168 h in a 5% CO2 incubator at 37°C, collected, and broken by three cycles of freezing (−80°C) and thawing. The culture medium was also collected. Cell lysates and the collected culture media were centrifuged at 7,000 × g for 10 min, and the obtained supernatants were used as virus stocks. The stock prepared from cell culture medium was used to infect the GEFs; eight passages were performed. The viral load of every passage was determined by qRT-PCR.
To determine viral growth characteristics, GEFs were seeded into 6-well plates and then infected with 20 μl of stock GEF-adapted GPgV (7th passage, 1.15 × 108 RNA copies/ml). At 12, 24, 36, 72, 96, 120, 144, 168, 192, and 216 h p.i., the cell culture supernatants and infected cells were collected. The cells were suspended in 200 μl of PBS. The GPgV genome copy numbers in these samples were determined.
To study in vitro coinfection of GPgV and GPV, GEFs were seeded into 6-well plates and then infected with GEF-adapted GPgV (7th passage, 1.15 × 108 RNA copies/ml) and GPV (6.46 × 106 DNA copies/ml). The culture medium and cell lysate, obtained using three freeze-thaw cycles, were collected at 168 h p.i., and GPgV genome copy numbers were determined.
To detect the host range of GPgV, DEFs, BHK-21, and Tb1Lu cells were seeded into 6-well plates and then infected with 200 μl of allantoic fluid from GPgV-infected goose embryos (14th passage, 5.83 × 109 RNA copies/ml); five blinded passages were performed. GPBLs and GSLs were isolated from adult geese using LTS1090G and LTS1090GPK reagents (TBD, China) and cultured in RPMI 1640 basic medium (Life Technologies, China) supplemented with 10% FBS at 37°C with 5% CO2. Cells grown in 12-well plates were infected with 200 μl of GEF-adapted GPgV (6th passage, 1.08 × 108 RNA copies/ml). Five blinded passages, each 48 h long, were performed. GPgV genome copy numbers in all obtained supernatants were determined.
In vivo experiments.
All goslings used for experimental inoculation were shown to be negative for known goose viruses. Three-day-old goslings were randomly divided into two groups: an experimental group (24 goslings) and a control group (20 goslings). Goslings in the experimental group were intravenously injected with 200 μl of allantoic fluid from GPgV-infected goose embryos (1.16 × 1010 RNA copies/ml); goslings in the control group were mock infected with 200 μl of PBS (Fig. 3A). The goslings were kept in a waterfowl isolator in separate rooms and observed daily for 30 days. Every gosling was weighed at PID 1, 4, 7, 10, 13, 16, 19, 22, 25, and 28. Dead goslings were necropsied immediately; the remaining goslings were sacrificed and necropsied at PID 30. Organs were sampled for HE staining, ICH, and for determination of viral loads.
In the GPgV/GPV coinfection experiment, 100 2-day-old GPV-positive goslings from GPV-positive breeding were randomly divided into an experimental group (54 goslings) and a control group (46 goslings) and were infected with GPgV or mock infected, respectively (Fig. 4A). The goslings were kept in a waterfowl isolator in separate rooms and observed daily for 21 days. Every gosling was weighed at PID 1, 4, 7, 10, 13, 16, 19, and 21. Dead goslings were necropsied immediately; the surviving goslings were sacrificed and necropsied at PID 21. Organs were sampled and used for analysis of viral (GPV and GPgV) loads.
Detection of GPgV negative-strand RNA by nested RT-PCR.
The assay was carried out as described previously (21). GPgV negative-strand RNA: GPgV-spreverse primers (Table 1) were used for the reverse transcription. Positive-sense RNA: a PCR amplified fragment using GPgV NF2 with T7 promoter TAATACGACTCACTATAGGGGGTCTACGCCTACAACCACT and GPgV R2 as primers. RNAs were transcribed from purified PCR products using a T7 mMESSAGE mMACHINE kit (Thermo Fisher Scientific, USA) according to the manufacturer’s protocols. In vitro-transcribed RNA is used for subsequent reverse transcription by the GPgV-spreverse primers (Table 1), which functioned as the negative control. H2O (no primer) used for a reverse transcription reaction functioned as the blank control. Then, GPgV NF2-R2 was used for the first round PCR and GPgV F6-R6 was used for the second round PCR (Table 1). RNA was extracted from the samples and the nested RT-PCR was performed as described above.
ACKNOWLEDGMENTS
This work was funded by grants from the National Key Research and Development Program of China (2017YFD0500800), Sichuan-International Joint Research for Science and Technology (2018HH0098), the China Agricultural Research System (CARS-42-17), and the Program Sichuan Veterinary Medicine and Drug Innovation Group of China Agricultural Research System (SCCXTD-2020-18).
REFERENCES
- 1.Stapleton JT, Foung S, Muerhoff AS, Bukh J, Simmonds P. 2011. The GB viruses: a review and proposed classification of GBV-A, GBV-C (HGV), and GBV-D in genus Pegivirus within the family Flaviviridae. J Gen Virol 92:233–246. doi: 10.1099/vir.0.027490-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kapoor A, Simmonds P, Scheel TK, Hjelle B, Cullen JM, Burbelo PD, Chauhan LV, Duraisamy R, Sanchez Leon M, Jain K, Vandegrift KJ, Calisher CH, Rice CM, Lipkin WI. 2013. Identification of rodent homologs of hepatitis C virus and pegiviruses. mBio 4:e00216-13–e00213. doi: 10.1128/mBio.00216-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith DB, Becher P, Bukh J, Gould EA, Meyers G, Monath T, Muerhoff AS, Pletnev A, Rico-Hesse R, Stapleton JT, Simmonds P. 2016. Proposed update to the taxonomy of the genera Hepacivirus and Pegivirus within the Flaviviridae family. J Gen Virol 97:2894–2907. doi: 10.1099/jgv.0.000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chandriani S, Skewes-Cox P, Zhong W, Ganem DE, Divers TJ, Van Blaricum AJ, Tennant BC, Kistler AL. 2013. Identification of a previously undescribed divergent virus from the Flaviviridae family in an outbreak of equine serum hepatitis. Proc Natl Acad Sci U S A 110:E1407–E1415. doi: 10.1073/pnas.1219217110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bailey AL, Buechler CR, Matson DR, Peterson EJ, Brunner KG, Mohns MS, Breitbach M, Stewart LM, Ericsen AJ, Newman CM, Koenig MR, Mohr E, Tan J, Capuano S, Simmons HA, Yang DT, O'Connor DH. 2017. Pegivirus avoids immune recognition but does not attenuate acute-phase disease in a macaque model of HIV infection. PLoS Pathog 13:e1006692. doi: 10.1371/journal.ppat.1006692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berg MG, Lee D, Coller K, Frankel M, Aronsohn A, Cheng K, Forberg K, Marcinkus M, Naccache SN, Dawson G, Brennan C, Jensen DM, Hackett J Jr, Chiu CY. 2015. Discovery of a novel human pegivirus in blood associated with hepatitis C virus co-infection. PLoS Pathog 11:e1005325. doi: 10.1371/journal.ppat.1005325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lozano G, Martínez-Salas E. 2015. Structural insights into viral IRES-dependent translation mechanisms. Curr Opin Virol 12:113–120. doi: 10.1016/j.coviro.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 8.Linnen J, Wages J Jr, Zhang-Keck ZY, Fry KE, Krawczynski KZ, Alter H, Koonin E, Gallagher M, Alter M, Hadziyannis S, Karayiannis P, Fung K, Nakatsuji Y, Shih JW, Young L, Piatak M Jr, Hoover C, Fernandez J, Chen S, Zou JC, Morris T, Hyams KC, Ismay S, Lifson JD, Hess G, Foung SK, Thomas H, Bradley D, Margolis H, Kim JP. 1996. Molecular cloning and disease association of hepatitis G virus: a transfusion-transmissible agent. Science 271:505–508. doi: 10.1126/science.271.5248.505. [DOI] [PubMed] [Google Scholar]
- 9.Baechlein C, Grundhoff A, Fischer N, Alawi M, Hoeltig D, Waldmann KH, Becher P. 2016. Pegivirus infection in domestic pigs, Germany. Emerg Infect Dis 22:1312–1314. doi: 10.3201/eid2207.160024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kapoor A, Simmonds P, Cullen JM, Scheel TK, Medina JL, Giannitti F, Nishiuchi E, Brock KV, Burbelo PD, Rice CM, Lipkin WI. 2013. Identification of a pegivirus (GB virus-like virus) that infects horses. J Virol 87:7185–7190. doi: 10.1128/JVI.00324-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simmonds P, Tuplin A, Evans DJ. 2004. Detection of genome-scale ordered RNA structure (GORS) in genomes of positive-stranded RNA viruses: implications for virus evolution and host persistence. RNA 10:1337–1351. doi: 10.1261/rna.7640104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis M, Sagan SM, Pezacki JP, Evans DJ, Simmonds P. 2008. Bioinformatic and physical characterizations of genome-scale ordered RNA structure in mammalian RNA viruses. J Virol 82:11824–11836. doi: 10.1128/JVI.01078-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simmonds P. 2012. SSE: a nucleotide and amino acid sequence analysis platform. BMC Res Notes 5:50. doi: 10.1186/1756-0500-5-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu G, Sun L, Ou J, Xu H, Wu L, Li S. 2018. Identification and genetic characterization of a novel parvovirus associated with serum hepatitis in horses in China. Emerg Microbes Infect 7:170. doi: 10.1038/s41426-018-0174-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang H, Wan Z, Xu R, Guan Y, Zhu N, Li J, Xie Z, Lu A, Zhang F, Fu Y, Tang S. 2017. A Novel human pegivirus HPgV-2 (HHpgV-1) is tightly associated with HCV infection and HCV/HIV-1 co-infection. Clin Infect Dis 66:29–35. doi: 10.1093/cid/cix748. [DOI] [PubMed] [Google Scholar]
- 16.Figueiredo AS, de Moraes MVDS, Soares CC, Chalhoub FLL, de Filippis AMB, Dos Santos DRL, de Almeida FQ, Godoi TLOS, de Souza AM, Burdman TR, de Lemos ERS, Dos Reis JKP, Cruz OG, Pinto MA. 2019. First description of Theiler's disease-associated virus infection and epidemiological investigation of equine pegivirus and equine hepacivirus coinfection in Brazil. Transbound Emerg Dis 66:1737–1751. doi: 10.1111/tbed.13210. [DOI] [PubMed] [Google Scholar]
- 17.Chu L, Jin M, Feng C, Wang X, Zhang D. 2019. A highly divergent hepacivirus-like flavivirus in domestic ducks. J Gen Virol 100:1234–1240. doi: 10.1099/jgv.0.001298. [DOI] [PubMed] [Google Scholar]
- 18.Quan PL, Firth C, Conte JM, Williams SH, Zambrana-Torrelio CM, Anthony SJ, Ellison JA, Gilbert AT, Kuzmin IV, Niezgoda M, Osinubi MO, Recuenco S, Markotter W, Breiman RF, Kalemba L, Malekani J, Lindblade KA, Rostal MK, Ojeda-Flores R, Suzan G, Davis LB, Blau DM, Ogunkoya AB, Alvarez Castillo DA, Moran D, Ngam S, Akaibe D, Agwanda B, Briese T, Epstein JH, Daszak P, Rupprecht CE, Holmes EC, Lipkin WI. 2013. Bats are a major natural reservoir for hepaciviruses and pegiviruses. Proc Natl Acad Sci U S A 110:8194–8199. doi: 10.1073/pnas.1303037110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li R, Li Y, Kristiansen K, Wang J. 2008. SOAP: short oligonucleotide alignment program. Bioinformatics 24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- 20.Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, Li Y, Li S, Shan G, Kristiansen K, Li S, Yang H, Wang J, Wang J. 2010. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res 20:265–272. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin L, Fevery J, Hiem Yap S. 2002. A novel strand-specific RT-PCR for detection of hepatitis C virus negative-strand RNA (replicative intermediate): evidence of absence or very low level of HCV replication in peripheral blood mononuclear cells. J Virol Methods 100:97–105. doi: 10.1016/S0166-0934(01)00399-8. [DOI] [PubMed] [Google Scholar]
- 22.Zhang J, Liu P, Wu Y, Wang M, Jia R, Zhu D, Liu M, Yang L, Yu Y, You Y, Chen S, Cheng A. 2019. Growth characteristics of the novel goose parvovirus SD15 strain in vitro. BMC Vet Res 15:63. doi: 10.1186/s12917-019-1807-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. 2001. Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol 146:2275–2289. doi: 10.1007/s007050170002. [DOI] [PubMed] [Google Scholar]