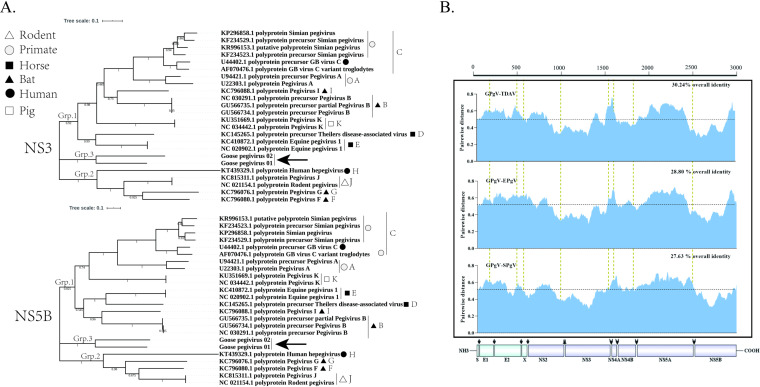
FIG 3.
Phylogenetic analysis of GPgV relative to other pegiviruses. Phylogenetic analysis was carried out using MEGA 7 software. (A) Phylogenetic trees of GPgV and representative strains from the Pegivirus genus built using the neighbor-joining method based on the sequences of NS3 (upper panel) and NS5B (lower panel) proteins. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (200 replicates) is shown next to the branches. The evolutionary distances were computed using the JTT matrix-based method and are presented in units of the number of amino acid substitutions per site. (B) Amino acid sequence divergence between polyproteins of GPgV, TDAV, EPgV, and SPgV. The sliding window size is 50 amino acid residues.