Contemporary ZIKV strains are closely related and often used interchangeably in laboratory research. Here, we identify two strain-specific determinants of ZIKV virulence that are evident in only Ifnar1−/− mice but not Ifnar1−/− Ifngr1−/− DKO mice. These results identify a balanced polymorphism in the E protein that is sufficient to attenuate the PRVABC59 strain but not H/PF/2013. We further identify a second virulence determinant in the H/PF/2013 strain, which is driven by the viral nucleotide sequence but not the amino acid sequence. Altogether, our work identifies a large and previously unreported difference in virulence between two commonly used ZIKV strains, in two widely used mouse models of ZIKV pathogenesis. Our results highlight that even very closely related virus strains can produce significantly different pathogenic phenotypes in common laboratory models.
KEYWORDS: IFNAR mouse, Zika virus, flavivirus, strain differences
ABSTRACT
Recent Zika virus (ZIKV) outbreaks and unexpected clinical manifestations of ZIKV infection have prompted an increase in ZIKV-related research. Here, we identify two strain-specific determinants of ZIKV virulence in mice. We found that strain H/PF/2013 caused 100% lethality in Ifnar1−/− mice, whereas PRVABC59 caused no lethality; both strains caused 100% lethality in Ifnar1−/− Ifngr1−/− double-knockout (DKO) mice. Deep sequencing revealed a high-frequency variant in PRVABC59 not present in H/PF/2013: a G-to-T change at nucleotide 1965 producing a Val-to-Leu substitution at position 330 of the viral envelope (E) protein. We show that the V330 variant is lethal on both virus strain backgrounds, whereas the L330 variant is attenuating only on the PRVABC59 background. These results identify a balanced polymorphism in the E protein that is sufficient to attenuate the PRVABC59 strain but not H/PF/2013. The consensus sequences of H/PF/2013 and PRVABC59 differ by 3 amino acids, but these were not responsible for the difference in virulence between the two strains. H/PF/2013 and PRVABC59 differ by an additional 31 noncoding or silent nucleotide changes. We made a panel of chimeric viruses with identical amino acid sequences but nucleotide sequences derived from H/PF/2013 or PRVABC59. We found that 6 nucleotide differences in the 3′ quarter of the H/PF/2013 genome were sufficient to confer virulence in Ifnar1−/− mice. Altogether, our work identifies a large and previously unreported difference in virulence between two commonly used ZIKV strains, in two widely used mouse models of ZIKV pathogenesis (Ifnar1−/− and Ifnar1−/− Ifngr1−/− DKO mice).
IMPORTANCE Contemporary ZIKV strains are closely related and often used interchangeably in laboratory research. Here, we identify two strain-specific determinants of ZIKV virulence that are evident in only Ifnar1−/− mice but not Ifnar1−/− Ifngr1−/− DKO mice. These results identify a balanced polymorphism in the E protein that is sufficient to attenuate the PRVABC59 strain but not H/PF/2013. We further identify a second virulence determinant in the H/PF/2013 strain, which is driven by the viral nucleotide sequence but not the amino acid sequence. Altogether, our work identifies a large and previously unreported difference in virulence between two commonly used ZIKV strains, in two widely used mouse models of ZIKV pathogenesis. Our results highlight that even very closely related virus strains can produce significantly different pathogenic phenotypes in common laboratory models.
INTRODUCTION
Zika virus (ZIKV) is a mosquito-transmitted flavivirus belonging to the Flaviviridae family of positive-sense single-stranded RNA viruses. ZIKV is related to other pathogenic flaviviruses, including dengue virus (DENV), yellow fever virus (YFV), West Nile virus (WNV), Japanese encephalitis virus (JEV), and tick-borne encephalitis virus (TBEV). Historically, ZIKV has been associated with a self-limiting rash and febrile illness that resolves within a few days, and these symptoms occur in only ∼20% of infected individuals (1, 2). However, new clinical manifestations associated with recent ZIKV outbreaks, including Guillain-Barré syndrome and congenital Zika syndrome, have stimulated a surge in ZIKV research (2–5).
Since the spread of ZIKV to the Western Hemisphere, numerous ZIKV strains have been isolated from patients and mosquitoes and distributed to laboratories across the world. Some widely used strains include FSS13025, which was isolated in 2010 from patient serum in Cambodia; H/PF/2013, which was isolated in 2013 from patient serum in French Polynesia; PRVABC59, which was isolated in 2015 from patient serum in Puerto Rico; Paraiba_01/2015, which was isolated in 2015 from patient serum in Brazil; and Mex-2-81, which was isolated in 2016 from an Aedes mosquito in Mexico (6–9). All of these strains share high nucleotide identity (at least 98.6% identity between any two strains) and minimal amino acid differences throughout their genomes. Thus, these closely related contemporary strains have been used largely interchangeably, and results from studies using various strains have been compared and combined to draw overall conclusions about the pathogenesis of contemporary ZIKV strains. Furthermore, various immunodeficient mouse models are used to study ZIKV pathogenesis, and the two most commonly used systems (Ifnar1−/− mice and Ifnar1−/− Ifngr1−/− double-knockout [DKO] mice) largely have been assumed to yield comparable results. Ifnar1−/− mice lack the type I interferon alpha/beta (IFN-α/β) receptor and are thus deficient in a key component of the innate antiviral response and highly susceptible to viral infections. Ifnar1−/− Ifngr1−/− DKO mice lack both the type I IFN-α/β receptor as well as the type II IFN-γ receptor and consequently are even more immunocompromised than Ifnar1−/− mice.
In this study, we identified two strain-specific determinants of ZIKV virulence that are evident in only Ifnar1−/− mice but not Ifnar1−/− Ifngr1−/− DKO mice. We identified a balanced polymorphism in the E protein that is sufficient to attenuate the PRVABC59 strain but not H/PF/2013. Furthermore, we identified a second virulence determinant in the H/PF/2013 strain that is driven by the viral nucleotide sequence and not the amino acid sequence.
RESULTS
ZIKV strains H/PF/2013 and PRVABC59 have distinct lethality phenotypes in Ifnar1−/− mice.
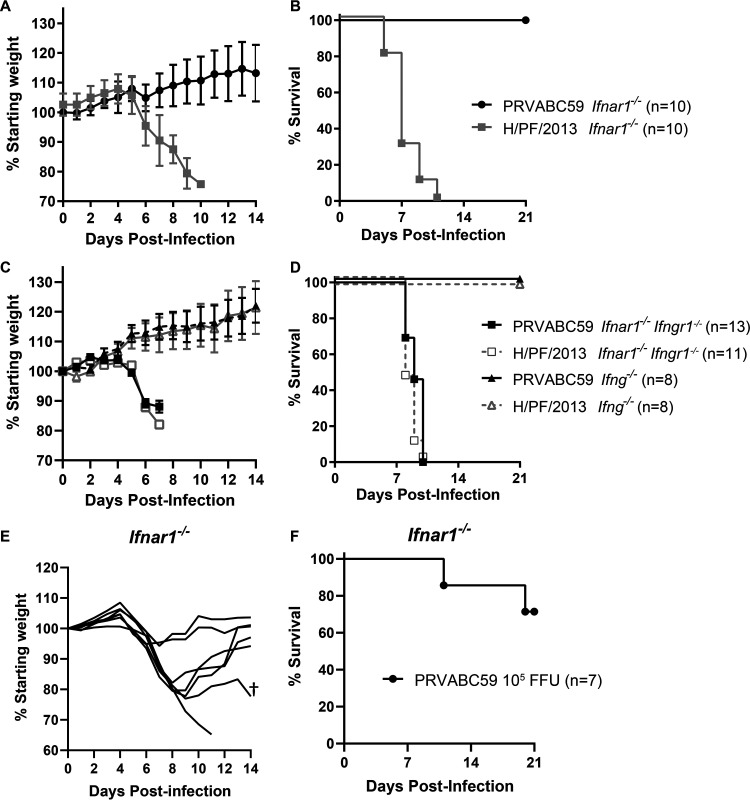
ZIKV strain H/PF/2013 was isolated from patient serum from a 2013 outbreak in French Polynesia, and ZIKV strain PRVABC59 was isolated from patient serum from a 2015 outbreak in Puerto Rico (7, 8). Deposited consensus sequences for these ZIKV strains (GenBank accession numbers KJ776791 and KU501215) as well as Sanger sequencing of our laboratory stocks of H/PF/2013 and PRVABC59 (10, 11) show that these two strains are very similar: they share >99% nucleotide identity (34 nucleotide differences across the genome) and have only 3 amino acid differences (1 in capsid and 2 in nonstructural protein 5 [NS5]). Both strains have been used widely and largely interchangeably by many research groups studying ZIKV virology and pathogenesis (10–19). However, we found that H/PF/2013 and PRVABC59 have distinct lethality phenotypes in Ifnar1−/− mice (Fig. 1). We infected 5- to 6-week-old Ifnar1−/− mice with 1 × 103 focus-forming units (FFU) of ZIKV H/PF/2013 or PRVABC59 by a subcutaneous route in the footpad and evaluated weight loss and lethality (Fig. 1A and B). These experiments used isolate virus stocks, originally derived from patient serum samples propagated through several passages on Vero cells. ZIKV H/PF/2013 was lethal in Ifnar1−/− mice, with mice beginning to lose weight at 6 days postinfection (dpi) and all mice succumbing by 11 dpi. In contrast, PRVABC59 was attenuated, causing no morbidity or mortality.
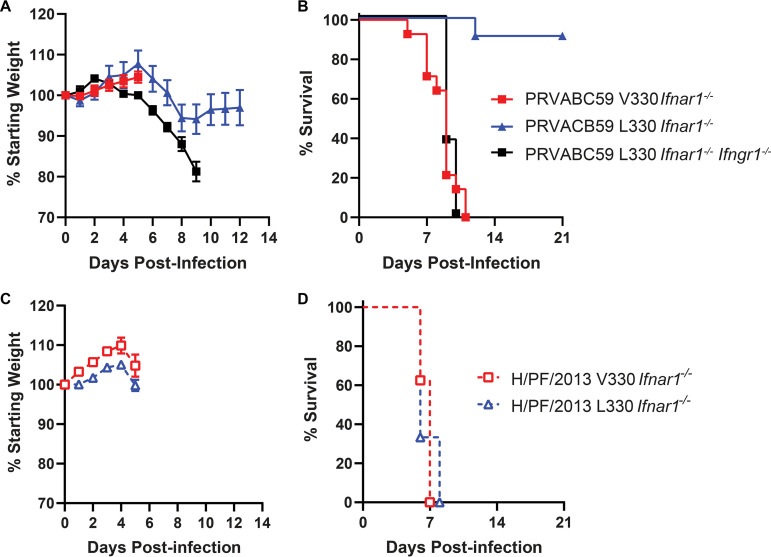
FIG 1.
PRVABC59 is attenuated in Ifnar1−/− mice. Five- to six-week-old Ifnar1−/−, Ifnar1−/− Ifngr1−/− DKO, or Ifng−/− mice were inoculated with 1 × 103 (A to D) or 1 × 105 (E and F) FFU of ZIKV strain PRVABC59 or H/PF/2013 isolate viruses by a subcutaneous route. Mice were weighed daily for 14 days, and weights are expressed as percentages of body weight prior to infection and were censored once one mouse in a group died. Results shown are the mean values ± standard errors of the means (SEM) for the indicated number of mice from one (E and F) or two (A to D) independent experiments. Lethality was monitored for 21 days. †, the mouse died at 20 dpi.
The attenuation of PRVABC59 was unexpected given its high similarity to H/PF/2013 and because other groups have shown the PRVABC59 isolate to be lethal in Ifnar1−/− Ifngr1−/− DKO mice (19, 20). To test this, we infected 5- to 6-week-old Ifnar1−/− Ifngr1−/− DKO mice, as well as Ifng−/− mice, with 1 × 103 FFU of PRVABC59 or H/PF/2013 isolate virus by a subcutaneous route in the footpad and evaluated weight loss and lethality (Fig. 1C and D). Ifnar1−/− × Ifngr1−/− mice began losing weight at 5 dpi, and all mice succumbed to either H/PF/2013 or PRVABC59 virus infection by 10 dpi. These results indicate that despite its attenuation in Ifnar1−/− mice, the PRVABC59 isolate can be lethal in a more susceptible mouse model (mice lacking both IFN-α/β and IFN-γ signaling). Unsurprisingly, since Ifng−/− mice retain IFN-α/β signaling, no Ifng−/− mice lost weight or succumbed to either virus, showing that the loss of IFN-γ signaling is not sufficient to render mice susceptible to ZIKV. To test whether PRVABC59 attenuation in Ifnar1−/− mice could be overcome with a higher inoculation dose, we infected 5- to 6-week-old Ifnar1−/− mice with 1 × 105 FFU of PRVABC59 isolate virus by a subcutaneous route in the footpad and evaluated weight loss and lethality (Fig. 1E and F). Mice began losing weight at 5 dpi, but 5 of 7 mice recovered. One mouse began losing weight at 5 dpi and continued to lose weight until it succumbed at 11 dpi, while another mouse lost weight from 5 to 9 dpi; began to recover, gaining weight until 16 dpi; and then succumbed at 20 dpi. Thus, even at a 100-fold-higher inoculation dose, the PRVABC59 isolate virus did not cause the ∼100% lethality characteristic of the H/PF/2013 strain in Ifnar1−/− mice. Altogether, these data reveal profoundly different virulence phenotypes between ZIKV strains PRVABC59 and H/PF/2013 in Ifnar1−/− mice despite their similar phenotypes in Ifnar1−/− Ifngr1−/− DKO mice. Given the high genetic similarity between PRVABC59 and H/PF/2013, we further investigated the viral determinants of these distinct virulence phenotypes.
The PRVABC59 isolate contains high-frequency variants.
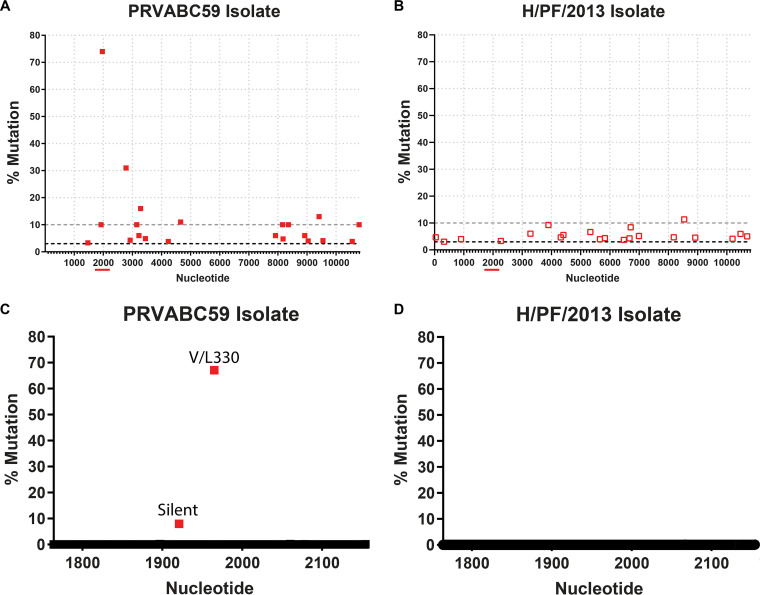
Although the consensus sequences of PRVABC59 and H/PF/2013 were highly similar, isolate viruses typically contain a mixture of genetic variants either present in the original patient sample or generated during passage in cell culture. To determine whether these two strains included distinct variants compared to their reference genomes, we sequenced RNA isolated from concentrated virus stocks using a deep-sequencing protocol. Most of the variants found in the two virus strains were present at relatively low frequencies (<10%), with the variants distributed across the genome. However, the PRVABC59 isolate contained 10 variants at a >10% frequency, whereas H/PF/2013 contained 1 variant at a >10% frequency, which was not shared between the two viruses (Fig. 2A and B). The PRVABC59 variant present at the highest frequency was a G-to-T mutation at nucleotide 1965, resulting in a Val-to-Leu substitution at amino acid position 330 in domain III of the envelope protein. These data are consistent with deep-sequencing results for the PRVABC59 strain from other groups, who also identified the V330L substitution (19–21), suggesting that this polymorphism is maintained through independent passages of this strain in separate laboratories. To quantify more accurately the frequency of G1965T variation in ZIKV stocks, we next used the Primer ID sequencing strategy. Primer ID is a deep-sequencing technique that incorporates an 11-nucleotide degenerative index into the cDNA synthesis primer, thereby uniquely identifying each template sequence and allowing resampled sequencing reads to be identified and pooled to create a highly accurate consensus sequence for each template (22). Primer ID confirmed an ∼70% frequency of the G1965T variant in the PRVABC59 isolate, whereas no variation was detected at position 1965 of strain H/PF/2013 (Fig. 2C and D). To investigate whether this variant was unique to ZIKV strain PRVABC59, we performed Primer ID on seven ZIKV strains propagated in either Vero or C6/36 cells, including two African-lineage strains (Dakar 41662 and Dakar 41671) and five Asian-lineage strains (H/PF/2013, PRVABC59, FSS13025, Paraiba_01/2015, and Mex-2-81). We also tested two related flaviviruses, Spondweni virus (SPOV) and DENV (Table 1). The published consensus sequences for all of these viruses encode a Val at position 330, except for SPOV, which encodes a Leu. We found that only the PRVABC59 strain has variation at position 1965 in virus grown in either Vero (69% T) or C6/36 (72% T) cells. These results indicate that the PRVABC59 isolate maintains T/G variation at position 1965 of the genome through multiple independent passages, suggesting a balanced polymorphism that is not found in other ZIKV strains or in either of two related flaviviruses. Next, we asked whether this variant was responsible for attenuating the PRVABC59 isolate in Ifnar1−/− mice.
FIG 2.
The PRVABC59 isolate contains high-frequency variants. (A and B) Deep sequencing was performed on ZIKV strains PRVABC59 and H/PF/2013. The cutoff for single nucleotide polymorphisms was set at a depth of ≥10 reads and a mutation rate of ≥3% (black dashed lines). The gray dashed line indicates a 10% mutation rate. (C and D) Primer ID was performed on ZIKV strains PRVABC59 and H/PF/2013 for the indicated 483-nucleotide region (red line under the full-genome sequence) covering amino acids 231 to 392 of the envelope protein.
TABLE 1.
Percent variation at nucleotide 1965 of viruses grown in Vero or C6/36 cells
| Virus | GenBank accession no. | Cell line | % T (Leu)a | % G (Val) residues |
|---|---|---|---|---|
| H/PF/2013 | KJ776791 | Vero | 0 | 100 |
| C6/36 | 0 | 100 | ||
| PRVABC59 | KU501215 | Vero | 69 | 31 |
| C6/36 | 72 | 28 | ||
| Paraiba | KX280026 | Vero | 0 | 100 |
| C6/36 | 0 | 100 | ||
| FSS13025 | KU955593 | Vero | 0 | 100 |
| C6/36 | 0 | 100 | ||
| Mex-2-81 | KX446950 | Vero | 0 | 100 |
| C6/36 | 0 | 100 | ||
| Dakar 41662 | KU955592.1 | Vero | 0 | 100 |
| C6/36 | 0 | 100 | ||
| Dakar 41671 | KU955595.1 | Vero | 0 | 100 |
| C6/36 | 0 | 100 | ||
| DENV4 | KU513442.1 | Vero | 0 | 100 |
| C6/36 | 0 | 100 | ||
| SPOV | KX227370.1 | Vero | 100 (CTG) | 0 |
| C6/36 | 100 (CTG) | 0 | ||
The Leu codon for ZIKV is TTA, and that for SPOV is CTG.
PRVABC59 V330 is lethal in Ifnar1−/− mice, whereas L330 is attenuated.
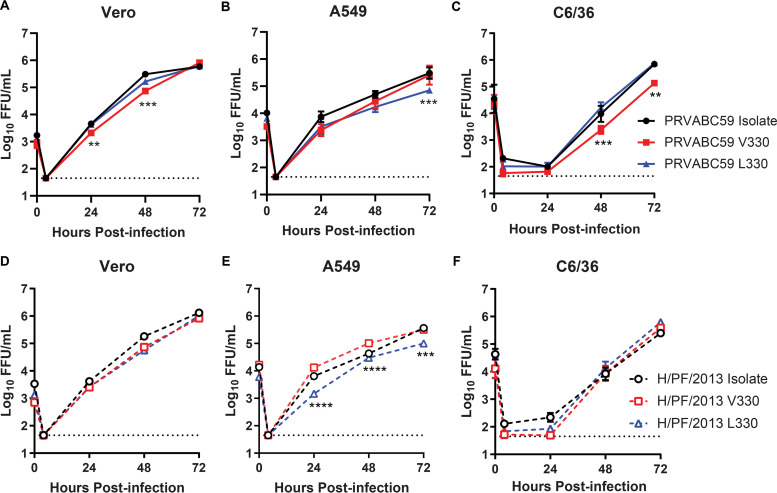
To test whether the G/T1965 variant affected ZIKV virulence, we generated infectious clones of PRVABC59 or H/PF/2013 with either a G at position 1965 (V330) or a T at position 1965 (L330) of the genome (Table 2). We evaluated the replication of the PRVABC59 and H/PF/2013 isolates, L330, and V330 viruses in Vero, A549, and C6/36 cells and did not find uniform differences (Fig. 3). Although there were statistically significant differences between the V330 and L330 viruses in some cell types, the magnitude of these differences was less than 10-fold, and the direction was inconsistent (PRVABC59 L330 greater than V330 in Vero and C6/36 cells; V330 greater than L330 in A549 cells for both backgrounds). These results indicate that V330L variation does not dramatically impact ZIKV replication in cell culture.
TABLE 2.
ZIKV infectious clones, mutants, and chimeras generated from the PRVABC59 or H/PF/2013 strain using a quadripartite infectious clone system
| Virus | Backbone fragmenta |
aa changeb,c (nt change) | % lethalityd | MTD (days)d | |||
|---|---|---|---|---|---|---|---|
| A | B | C | D | ||||
| H/PF/2013 V330 | PF | PF | PF | PF | Consensus | 100 | 6.6 |
| H/PF/2013 L330 | PF | PF | PF | PF | V330L (G1965T) | 100 | 6.7 |
| PRVABC59 V330 | PRV | PRV | PRV | PRV | Consensus | 100 | 8.6 |
| PRVABC59 L330 | PRV | PRV | PRV | PRV | V330L (G1965T) | 10 | 15.0 |
| V020 | PRV | PRV | PRV | PRV | I75T (C346T) | 0 | None |
| V330L (G1965T) | |||||||
| V37A (T7939C) | |||||||
| V60M (G8007A) | |||||||
| V024 | PRV | PRV | PF | PF | I75T (C346T) | 70 | 12.4 |
| V330L (G1965T) | |||||||
| V025 | PF | PF | PRV | PRV | V330L (G1965T) | 0 | None |
| V37A (T7939C) | |||||||
| V60M (G8007A) | |||||||
| V026 | PRV | PRV | PF | PRV | I75T (C346T) | 20 | 13.5 |
| V330L (G1965T) | |||||||
| V027 | PRV | PRV | PRV | PF | I75T (C346T) | 100 | 9.4 |
| V330L (G1965T) | |||||||
| V37A (T7939C) | |||||||
| V60M (G8007A) | |||||||
PRV, PRVABC59; PF, H/PF/2013.
Amino acid (aa) numbering is for individual proteins (C, E, or NS5), and nucleotide (nt) numbering is for the complete genome.
I75T is in C, V330L is in E, and V2611A and V2634M are in NS5.
Percent lethality and mean time to death (MTD) determined in Ifnar1−/− mice.
FIG 3.
The V330L substitution does not uniformly impact ZIKV replication in cell culture. Vero (A and D), A549 (B and E), or C6/36 (C and F) cells were infected with ZIKV PRVABC59 or H/PF/2013 isolate, V330 clone, or L330 clone virus at an MOI of 0.01. Viruses in culture supernatants were titrated by a focus-forming assay. Data shown are the mean values ± SEM for 9 samples from 3 independent experiments. V330 and L330 viruses were compared to each other by 2-way ANOVA with multiple comparisons. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
To evaluate virulence, we infected 5- to 6-week-old Ifnar1−/− mice with 1 × 103 FFU of PRVABC59 or H/PF/2013 V330 or L330 clone virus by a subcutaneous route in the footpad and evaluated weight loss and lethality (Fig. 4). Mice infected with PRVABC59 L330 lost weight from 6 to 9 dpi, with only 1 of 7 mice succumbing, whereas mice infected with PRVABC59 V330 virus began losing weight at 6 dpi, and all mice succumbed by 11 dpi (Fig. 4A and B). All mice infected with either the H/PF/2013 V330 or L330 virus began losing weight at 6 dpi and succumbed by 8 dpi (Fig. 4C and D). To determine whether the PRVABC59 L330 virus could be lethal in a more susceptible mouse model, we infected 8-week-old Ifnar1−/− Ifngr1−/− DKO mice with 1 × 103 FFU by a subcutaneous route in the footpad. Mice began to lose weight at 4 dpi, and all mice succumbed by 10 dpi (Fig. 4A and B). These results show that the L330 variant is attenuating on the PRVABC59 genetic background (with a phenotype similar to that of the PRVABC59 isolate virus) but did not impact virulence on the H/PF/2013 genetic background, altogether implying an epistatic effect of L330 on virulence.
FIG 4.
The V330L substitution is attenuating on a PRVABC59 but not on an H/PF/2013 background in Ifnar1−/− mice. Five- to six-week-old Ifnar1−/− mice or 8-week-old Ifnar1−/− Ifngr1−/− DKO mice were inoculated with 1 × 103 FFU of ZIKV strain PRVABC59 or H/PF/2013 V330 or L330 viruses by a subcutaneous route. (A and C) Mice were weighed daily for 14 days. Weights are expressed as percentages of body weight prior to infection and were censored once one mouse in a group died. (B and D) Lethality was monitored for 21 days. Results shown are the means ± SEM for 11 to 14 Ifnar1−/− mice (PRVABC59 V330 and L330) and 9 Ifnar1−/− mice (H/PF/2013 V330 and L330) from two independent experiments or 8 Ifnar1−/− Ifngr1−/− DKO mice from one experiment.
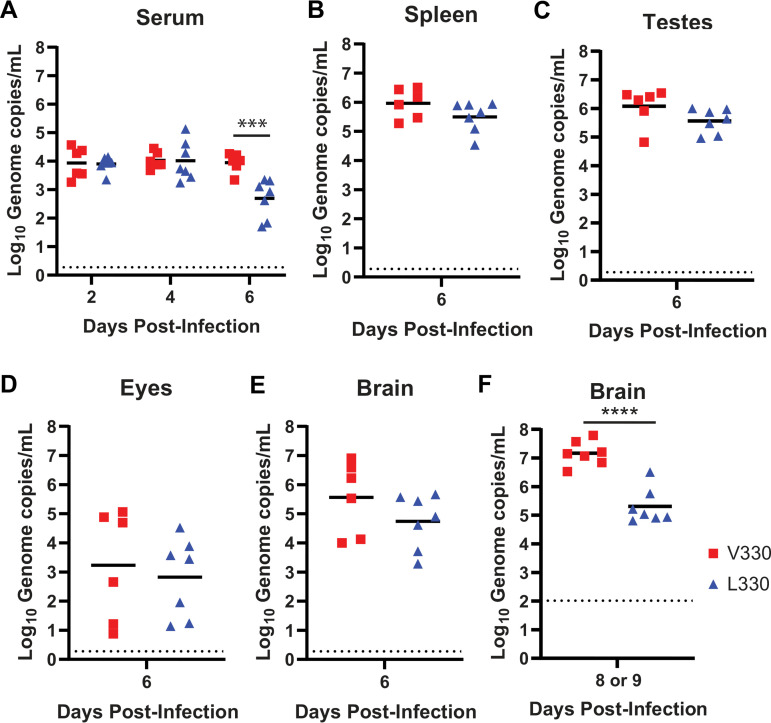
Since the PRVABC59 V330 and L330 viruses produced distinct lethality phenotypes in Ifnar1−/− mice, we next compared viral loads in serum and tissues (Fig. 5). We infected 5- to 6-week-old Ifnar1−/− mice with 1 × 103 FFU of the PRVABC59 V330 or L330 virus by a subcutaneous route in the footpad and measured viral loads in serum at 2, 4, and 6 dpi (Fig. 5A) and viral loads in tissues (spleen, brain, eyes, and testes) at 6 dpi by quantitative reverse transcription-PCR (qRT-PCR) (Fig. 5B to E). Compared to the V330 virus, the L330 virus produced similar viral loads at 2 and 4 dpi but significantly lower viral loads at 6 dpi in serum (10-fold reduction). There was no significant difference between the PRVABC59 V330 and L330 viral loads in any tissues tested at 6 dpi. However, at 6 dpi, mice infected with either the V330 or L330 virus exhibited minimal morbidity. To evaluate viral loads at a time point with a distinct disease presentation, we infected Ifnar1−/− mice as described above but harvested tissues at 8 or 9 dpi, when all V330-infected mice exhibited disease signs constituting a humane endpoint and L330-infected mice exhibited no overt disease signs. One PRVABC59 V330-infected mouse was moribund and harvested at 8 dpi, and all other V330-infected mice were moribund and harvested at 9 dpi, along with all L330-infected mice. We measured viral loads in the brain by qRT-PCR (Fig. 5F). Mice infected with the PRVABC59 L330 virus had significantly lower viral loads in the brain than did V330-infected mice at 8 to 9 dpi. Sanger sequencing of viruses isolated from the brains of infected mice confirmed that the viruses maintained their original V330 or L330 genotype, with no additional mutations in E. The lower viral loads in the brains of L330-infected mice are consistent with the attenuated phenotype of this virus and, combined with lower viremia at 6 dpi, suggest a more efficient clearance of the L330 virus than of V330. We considered whether a higher genome/FFU ratio for the L330 virus might result in enhanced immune stimulation and more rapid clearance. However, we found that the genome/FFU ratio of the PRVABC59 L330 virus stock was similar to that of the V330 virus (213 versus 450).
FIG 5.
PRVABC59 L330 produces low viral loads late in infection. Five- to six-week-old Ifnar1−/− mice were inoculated with 1 × 103 FFU of ZIKV strain PRVABC59 V330 or L330 virus by a subcutaneous route in the footpad. (A) Blood was collected 2, 4, and 6 days after infection, and ZIKV RNA in serum was measured by qRT-PCR. (B to E) Mice were euthanized 6 days after infection and perfused, and tissues were harvested. ZIKV RNA in tissue was measured by qRT-PCR. (F) V330-infected mice were euthanized when they reached a humane endpoint (8 or 9 dpi) and L330-infected mice were euthanized at 9 dpi, tissues were harvested, and ZIKV RNA in the brain was measured by qRT-PCR. ****, P < 0.0001 (unpaired 2-tailed t test). Results shown represent data from 2 independent experiments.
Nucleotide differences near the 3′ end of the H/PF/2013 genome confer lethality.
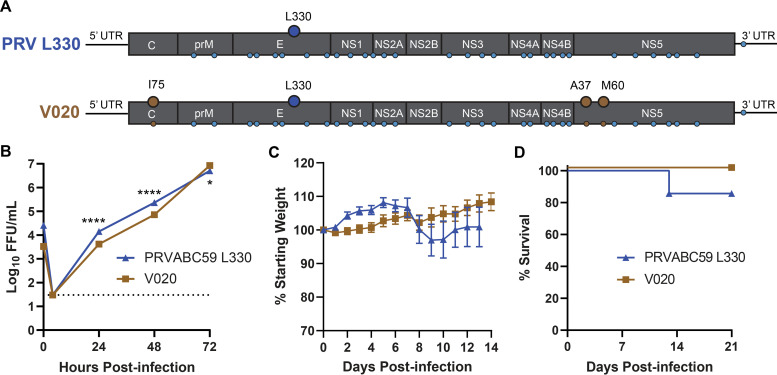
Since the variation at position 330 of the envelope protein affected virulence only on the PRVABC59 background and not on the H/PF/2013 background, other differences between the two viruses must contribute to virulence in Ifnar1−/− mice. The consensus sequences of PRVABC59 and H/PF/2013 differ by only 34 nucleotides throughout the genome. These include 30 silent changes, 1 noncoding change, and 3 coding changes (one in capsid [I/T75] and two in the methyltransferase domain of NS5 [V/A37 and V/M60]). To test whether any of the 3 amino acids that differ between the two viruses were responsible for their differential lethality in Ifnar1−/− mice, we used site-directed mutagenesis to generate a PRVABC59 L330 virus (V020) with an amino acid sequence identical to that of H/PF/2013 L330 but maintaining the 31 silent and noncoding nucleotides from the PRVABC59 genome (Table 2 and Fig. 6A). We compared the replication of V020 to that of PRVABC59 L330 in Vero cells, and while there were statistically significant differences in viral titers, the magnitude of the difference was small (greatest difference, <4-fold). Thus, we concluded that V020 was not markedly impaired in replication in Vero cells (Fig. 6B). We infected 5- to 6-week-old Ifnar1−/− mice with 1 × 103 FFU of PRVABC59 L330 or V020 by a subcutaneous route in the footpad and evaluated weight loss and lethality (Fig. 6C and D). Mice infected with PRVABC59 L330 began losing weight from 7 to 10 dpi, with only 1 of 7 mice succumbing to the infection, similar to previous experiments (Fig. 4). Mice infected with V020 did not lose weight, and no mice succumbed to the infection. These results indicate that the 3 amino acids that differ between the consensus sequences of the PRVABC59 and H/PF/2013 viruses are not responsible for their differential lethality in Ifnar1−/− mice.
FIG 6.
Three amino acids that differ between H/PF/2013 and PRVABC59 do not confer lethality. (A) Schematic of the PRVABC59 L330 and V020 genomes depicting the introduction of 3 H/PF/2013 amino acids (I75 in C and A37 and M60 in NS5) on the PRVABC59 L330 background; small circles indicate nucleotide differences between PRVABC59 and H/PF/2013. (B) Vero cells were infected with ZIKV PRVABC59 L330 or V020 at an MOI of 0.01. Viruses in culture supernatants were titrated by a focus-forming assay. Data shown are the means ± SEM for 9 samples from 3 independent experiments. *, P < 0.05; ****, P < 0.0001 (2-way ANOVA with multiple comparisons). (C and D) Five- to six-week-old Ifnar1−/− mice were inoculated with 1 × 103 FFU of ZIKV strain PRVABC59 L330 or V020 virus by a subcutaneous route. Mice were weighed daily for 14 days, and weights are expressed as percentages of body weight prior to infection and were censored once one mouse in a group died. Results shown are the mean values ± SEM for 7 mice for PRVABC59 L330 and 11 mice for V020 from four independent experiments. Lethality was monitored for 21 days.
Since amino acid changes did not explain the observed differences in virulence between the PRVABC59 L330 and H/PF/2013 L330 viruses, we next investigated whether the remaining 31 nucleotide differences between the two strains were responsible. The infectious clone system used to generate PRVABC59 and H/PF/2013 viruses divides the viral genome over four plasmids (fragments A, B, C, and D) (10, 11). The high sequence identity between PRVABC59 and H/PF/2013 preserves the restriction sites used to ligate each genomic fragment, making it straightforward to generate chimeras of the two strains. We generated chimeric viruses encoding the PRVABC59 L330 amino acid sequence but the nucleotide sequence of either PRVABC59 or H/PF/2013 and evaluated their virulence in Ifnar1−/− mice (Table 2 and Fig. 7A). All chimeric viruses tested replicated equivalently to the wild-type (WT) L330 virus in Vero cells, except for V024, which had significantly lower titers at 24 or 48 h (Fig. 7B). We first tested viruses with the 5′ half (fragments A and B) derived from the PRVABC59 nucleotide sequence and the 3′ half (fragments C and D) derived from the H/PF/2013 nucleotide sequence (V024) or vice versa (V025) (Fig. 7A). We infected 5- to 6-week-old Ifnar1−/− mice with 1 × 103 FFU of V024 or V025 virus by a subcutaneous route in the footpad and evaluated weight loss and lethality (Fig. 7C and D). Mice infected with V024 began losing weight at 7 dpi, and 6 of 8 mice succumbed to the virus by 11 dpi, whereas mice infected with V025 did not lose weight, and no mice succumbed to the virus. Since both viruses encoded the same amino acid sequence, these data indicate that nucleotides on the 3′ half of the H/PF/2013 genome confer virulence in Ifnar1−/− mice. To further define which of these 19 nucleotide differences contribute to the virulent phenotype, we generated chimeric viruses still encoding the PRVABC59 L330 amino acid sequence but with either the C fragment (V026) or D fragment (V027) having the nucleotide sequence of H/PF/2013. The C fragment contains 13 nucleotide differences between the two strains, and the D fragment contains 6. We infected 5- to 6-week-old Ifnar1−/− mice with 1 × 103 FFU of V026 or V027 virus by a subcutaneous route in the footpad and evaluated weight loss and lethality (Fig. 7E and F). Mice infected with V026 lost weight from 6 to 12 dpi, with 7 of 9 mice recovering, whereas mice infected with V027 began losing weight at 6 dpi, and all mice succumbed to the virus by 11 dpi. To test if the increased virulence of V027 corresponded to higher viral loads, we infected 5- to 6-week-old Ifnar1−/− mice with 1 × 103 FFU of V026 or V027 virus by a subcutaneous route in the footpad and harvested tissues to measure ZIKV RNA by qRT-PCR (Fig. 7G). V027-infected mice were euthanized at 9 dpi, when the mice exhibited disease signs constituting a humane endpoint. Tissues from V026-infected mice were harvested at the same time, although these mice did not exhibit severe disease signs. Brain viral loads in V027-infected mice were significantly higher (10-fold) than those in V026-infected mice at 9 dpi, consistent with the greater virulence of the V027 virus. Sanger sequencing of viruses isolated from the brains of infected mice confirmed that the viruses maintained the 6 nucleotides that differ in the D fragment, with no additional mutations. These data suggest that the 6 nucleotides that differ between the two viruses on the D fragment are sufficient to confer the virulent phenotype of the H/PF/2013 strain.
FIG 7.
Nucleotides at the 3′ end of ZIKV strain H/PF/2013 confer lethality in mice. (A) Schematic of PRVABC59 and H/PF/2013 chimeric viruses (V024 to V027). Brown circles depict the introduction of 3 H/PF/2013 amino acids; small circles indicate nucleotide differences between PRVABC59 and H/PF/2013. (B) Vero cells were infected with ZIKV H/PF/2013 L330, V024, V025, V026, or V027 at an MOI of 0.01. Viruses in culture supernatants were titrated by a focus-forming assay. Data shown are the means ± SEM for 9 samples from 3 independent experiments. **, P < 0.01; ***, P < 0.001 (2-way ANOVA). (C to G) Five- to six-week-old Ifnar1−/− mice were inoculated with 1 × 103 FFU of ZIKV V024, V025, V026, or V027 by a subcutaneous route. Mice were weighed daily for 21 days, and weights are expressed as percentages of body weight prior to infection and were censored once one mouse in a group died. Results shown are the mean values ± SEM for the indicated numbers of Ifnar1−/− mice for each virus from three independent experiments. Lethality was monitored for 21 days. (G) Mice were euthanized 9 days after infection, and tissues were harvested. ZIKV RNA in the brain was measured by qRT-PCR. *, P < 0.05 (unpaired 2-tailed t test).
Altogether, this work identifies a large and previously unreported difference in virulence between two commonly used ZIKV strains. Our data further define the effect of a previously reported variant in the PRVABC59 strain. We also identify a novel role for noncoding changes as ZIKV virulence determinants, suggesting possible effects on RNA properties such as RNA structure or posttranscriptional modifications.
DISCUSSION
The new clinical manifestations associated with ZIKV after its emergence in the Western Hemisphere, particularly congenital Zika syndrome, led to a surge in ZIKV research. Many different ZIKV strains have been isolated during recent outbreaks and distributed to research laboratories around the world (e.g., ∼12 contemporary strains available from BEI Resources and ∼20 from the World Reference Center for Emerging Viruses and Arboviruses). All contemporary ZIKV strains share high nucleotide identity and minimal amino acid differences throughout their genomes and thus have been used largely interchangeably to draw conclusions about the pathogenesis of contemporary ZIKV strains. However, not all contemporary ZIKV strains share the same pathogenic outcome in mice. Both a ZIKV strain isolated in 2013 from French Polynesia (H/PF/2013) and a strain isolated in 2015 from Brazil (Paraiba_01/2015) have been shown to cause 100% lethality in Ifnar1−/− mice (23, 24). In contrast, a ZIKV strain isolated in 2010 from Cambodia (FSS13025) was only 20 to 30% lethal in Stat2−/− or Ifnar1−/− mice, and a strain isolated in 2015 from Puerto Rico (PRVABC59) was not lethal in either mouse model (25). However, the H/PF/2013, FSS13025, and PRVABC59 strains were uniformly lethal in Ifnar1−/− Ifngr1−/− DKO mice (19, 20, 26, 27). These data indicate ZIKV strain-specific contributions to virulence in mice and highlight that differences in pathogenesis may not be evident in highly susceptible models, such as Ifnar1−/− Ifngr1−/− DKO mice, or in younger mice. Strain characteristics such as virus source (human versus mosquito), disease outcome, geographic origin, or year of isolation do not obviously correlate with the pathogenic phenotype in mice, although such relationships might be revealed by comparing a larger number of strains under identical experimental conditions (23, 25, 28). ZIKV lethality is also age dependent, with younger mice being more susceptible than older mice (29). Accordingly, Manangeeswaran et al. found that neonatal (10-day-old) Ifnar1−/− mice were susceptible to the PRVABC59 isolate (12), whereas the 5- to 6-week old Ifnar1−/− mice used in our experiments were not. In this study, we describe two distinct genetic determinants of ZIKV strain-specific virulence; notably, both phenotypes were evident in Ifnar1−/− mice but masked in Ifnar1−/− Ifngr1−/− DKO mice. Our results further define the effect of a previously reported variant in the PRVABC59 strain (19), identify a novel role for noncoding changes as ZIKV virulence determinants, and highlight that even very closely related virus strains can produce significantly different pathogenic phenotypes in common laboratory models.
Duggal et al. previously reported variants in the PRVABC59 isolate virus that led to attenuated pathogenesis in Ifnar1−/− Ifngr1−/− DKO mice (19). Their deep sequencing revealed the presence of a Val-to-Leu substitution at position 330 of the E protein as well as a Trp-to-Gly substitution at position 98 of nonstructural protein 1 (NS1). Those authors concluded that the substitutions were tissue culture adaptations, as these variants were found at very low levels upon deep sequencing of the original patient sera and were found to increase in frequency over three passages in Vero cells. However, while these substitutions increased in frequency with passage, it is not clear whether they are truly adaptive. Notably, we did not detect the L330 substitution in the H/PF/2013 strain, even though our stock of this strain has been passaged more extensively in Vero cells than the PRVABC59 strain, nor was this substitution detected in other ZIKV strains that we tested (although many of these have less certain passage histories). Furthermore, the frequency of the V330 and L330 variants remains constant (∼70% L330) in independent passages of the PRVABC59 strain grown on different cell types (Vero versus C6/36) and by different laboratories. The stability of this variation could suggest a balanced polymorphism at this site, although the selective advantage of this or why it would be advantageous only on the PRVABC59 genetic background is unclear.
Duggal et al. found that the L330 substitution attenuated the virus in 12- to 16-week-old Ifnar1−/− Ifngr1−/− DKO mice, increasing the mean time to death, and the addition of the NS1 G98 substitution further attenuated the virus, highlighting the ability of this variant to act epistatically to modulate virulence. We also identified the E L330 and NS1 G98 variants in our PRVABC59 isolate, which were not present in our H/PF/2013 stock. We found that the L330 substitution was sufficient to attenuate the PRVABC59 virus in Ifnar1−/− mice (recapitulating the phenotype of the isolate virus), so we did not further investigate the NS1 G98 variant.
ZIKV replicates poorly in mice with intact IFN-α/β signaling due to an inability to antagonize murine STAT2 and STING (30–33). Thus, mouse pathogenesis studies typically use mice deficient in IFN-α/β signaling, and both Ifnar1−/− and Ifnar1−/− Ifngr1−/− DKO mice are widely used in the field (20, 23, 26, 27, 34, 35). However, Ifnar1−/− Ifngr1−/− DKO mice develop severe disease and succumb rapidly to ZIKV infection, independent of the inoculation route, compared to Ifnar1−/− mice (26, 29). Consistent with the phenotype of the PRVABC59 isolate virus, our PRVABC59 L330 virus caused 100% lethality in Ifnar1−/− Ifngr1−/− DKO mice but only 10% lethality in Ifnar1−/− mice.
We found that the L330 substitution was sufficient to attenuate virulence on the PRVABC59 genetic backbone (10% lethality with PRVABC59 L330, compared to 100% lethality with PRVABC59 V330 in Ifnar1−/− mice). So it was surprising that this same substitution had no effect on the H/PF/2013 backbone (the L330 virus remained 100% lethal). The E protein facilitates membrane fusion between the virus and the host cell and is the primary viral protein against which neutralizing antibodies are produced (36–38). Flavivirus E proteins are class II fusion proteins with three distinct domains: domain I (EDI), domain II (EDII), and domain III (EDIII) (39). EDI and EDII are discontinuous, forming a central β-barrel and an elongated dimerization region, respectively, while EDIII forms a continuous C-terminal, immunoglobulin-like domain. It is not obvious how a Val-to-Leu substitution in EDIII would attenuate virulence. Compared to the V330 virus, PRVABC59 L330 produced low viral loads in the serum at 6 dpi and in the brain at 9 dpi, despite similar viral loads earlier in infection, suggesting that the L330 virus may be cleared more efficiently. However, why this would be specific to the PRVABC59 strain remains unclear. Apart from the V330L variation, the PRVABC59 and H/PF/2013 strains have identical E protein amino acid sequences, so the L330 substitution would be expected to have a similar effect on both strains. An alternative explanation is that the distinct virulence phenotypes are determined not by the V330L change in the E protein but by the effect of the G1965T nucleotide change on RNA structure or other RNA functions. Unfortunately, the codons in this position preclude the generation of mutants that would distinguish the effects of nucleotide and amino acid changes at this site.
Since the L330 substitution was not attenuating on the H/PF/2013 background, and the other 3 amino acids that differ between the two strains did not have an effect on virulence, we sought to identify which of the other 31 nucleotide differences between the two strains were responsible for the differential lethality. Using chimeric viruses, we were able to narrow down the virulence determinant to 6 nucleotide differences in the 3′ region of the H/PF/2013 genome (including NS5 and the 3′ untranslated region [UTR]), although the mechanism by which these nucleotide changes modulate virulence remains unclear. Possible mechanisms could include effects on RNA structure and/or posttranscriptional modifications, although according to a Web server for predicted RNA secondary structure (https://rna.urmc.rochester.edu/RNAstructureWeb/Servers/Fold/Fold.html), these changes did not obviously disrupt predicted RNA structures. These changes also did not correspond to reported long-range RNA interactions within the ZIKV genome (40, 41) or experimentally determined m6A modification sites (42). One of the nucleotide differences falls 6 nucleotides upstream of the first stem-loop structure in the 3′ UTR. Thus, it is plausible that one or more of the nucleotide changes between PRVABC59 and H/PF/2013 could impact the production, stability, or function of subgenomic flavivirus RNA (sfRNA). sfRNA is a stable RNA degradation product resulting from the stalling of the cellular 5′-to-3′ exoribonuclease Xrn1 on RNA structures in the flavivirus 3′ UTR, which has been shown to modulate the replication and virulence of ZIKV and other flaviviruses (43, 44).
Altogether, our work identifies a large and previously unreported difference in virulence between two commonly used ZIKV strains, in two widely used mouse models of ZIKV pathogenesis (Ifnar1−/− and Ifnar1−/− Ifngr1−/− DKO mice). Our data further describe the effect of a previously reported variant in the PRVABC59 strain that appears to be attenuating only on the PRVABC59 genetic background. We also identified a novel role for noncoding changes as ZIKV virulence determinants, suggesting possible effects on RNA structure or posttranscriptional modifications. Our results highlight that even very closely related virus strains can produce significantly different pathogenic phenotypes in common laboratory models.
MATERIALS AND METHODS
Viruses and cells.
ZIKV strains H/PF/2013 and PRVABC59 were provided by the U.S. Centers for Disease Control and Prevention (8, 45). ZIKV strains Dakar 41662, Dakar 41671, FSS13025, and Mex-2-81, as well as SPOV, were provided by the World Reference Center for Emerging Viruses and Arboviruses (6–8). ZIKV strain Paraiba_01/2015 was obtained from Michael Diamond (Washington University in St. Louis), and DENV4 was obtained from Aravinda de Silva (UNC) (9, 46). Virus stocks were grown in Vero (African green monkey kidney epithelial) cells. Titers of virus stocks were determined on Vero cells by a focus-forming assay (FFA) (47). Vero and A549 cells were maintained in Dulbecco’s modified Eagle medium (DMEM) containing 5% heat-inactivated fetal bovine serum (FBS) and l-glutamine at 37°C with 5% CO2. C6/36 cells were maintained in DMEM containing 6% FBS, nonessential amino acids (NEAA), and penicillin-streptomycin (P/S) at 32°C with 5% CO2. For multistep growth analysis, cells were infected at a multiplicity of infection (MOI) of 0.01 and incubated at 37°C or 32°C with 5% CO2 for 1 h. Next, the inoculum was aspirated, cells were washed with phosphate-buffered saline (PBS), and the medium was replenished. Samples of the infected cell culture supernatant were collected at 4, 24, 48, and 72 h postinfection and stored at −80°C for virus titration. Virus quantification was performed by an FFA on Vero cells. Duplicates of serial 10-fold dilutions of virus in viral growth medium (DMEM containing 2% FBS and 20 mM HEPES) were applied to Vero cells in 96-well plates and incubated as described above for 1 h. Following virus adsorption, the monolayers were overlaid with 1% methylcellulose in Eagle minimum essential medium (MEM). Infected cell foci were detected 42 to 46 h after infection. Following fixation with 2% paraformaldehyde for 1 h at room temperate, plates were incubated with 500 ng/ml of flavivirus-cross-reactive mouse monoclonal antibody (MAb) E60 (48) for 2 h at room temperature or overnight at 4°C. After incubation at room temperature for 2 h with a 1:5,000 dilution of horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (Sigma), foci were detected by the addition of the TrueBlue substrate (KPL). Foci were quantified with an Immunospot analyzer (Cellular Technology Limited).
ZIKV infectious clone mutagenesis.
We used previously described infectious clones of ZIKV strain H/PF/2013 or PRVABC59, generated using a quadripartite infectious clone system (10, 11). Due to the high sequence identity between H/PF/2013 and PRVABC59, the same naturally occurring class IIG nonpalindromic restriction endonuclease sites within the full-length genome were used for both infectious clones, allowing us to swap fragments between the two strains to generate chimeric viruses. PCR site-directed mutagenesis was used to introduce mutations into H/PF/2013 or PRVABC59 plasmids. The resulting purified plasmids were digested, ligated, in vitro transcribed, and electroporated into Vero or C6/36 cells as previously described (49). Supernatants from electroporated Vero or C6/36 cells were harvested after 6 to 7 days and passaged once on Vero cells to generate virus stocks. Virus stocks were titrated by an FFA on Vero cells. Restriction enzymes and the Phusion high-fidelity PCR kit were obtained from New England BioLabs. A SuperScript III first-strand synthesis kit was obtained from Invitrogen. Oligonucleotide primers and probes for DNA amplification and qRT-PCR and sequencing primers were obtained from Sigma and IDT. The mMachine T7 Ultra transcription kit was obtained from Ambion. Antiflavivirus monoclonal antibody E60 was produced by the UNC Protein Expression and Purification Core Facility (48). Secondary antibodies were obtained from Sigma.
Mouse experiments.
Animal husbandry and experiments were performed under the approval of the University of North Carolina at Chapel Hill Institutional Animal Care and Use Committee. Five- to six-week-old, or 8-week-old, male and female Ifnar1−/−, Ifnar1−/− Ifngr1−/− DKO, or Ifng−/− mice on a C57BL/6 background were used. Mice were inoculated with 1 × 103 or 1 × 105 FFU of ZIKV in a volume of 50 μl by a subcutaneous route in the footpad. Survival and weight loss were monitored for 14 or 21 days. Animals that lost ≥30% of their starting weight or that exhibited severe disease signs were euthanized. Weights are reported as percentages of weights on the day of infection, and group means were censored once one animal in a group died.
Measurement of viral burden.
ZIKV-infected mice were sacrificed at 6 to 9 dpi and perfused with 20 ml of PBS. Spleen, kidney, testes, brain, and eyes were harvested and homogenized with zirconia beads (BioSpec) in a MagNA Lyser instrument (Roche Life Science) in 500 μl (eyes) or 1 ml (all other tissues) of RLT buffer (Qiagen). Blood was collected at 2 and 4 dpi by submandibular bleed with a 5-mm Goldenrod lancet and by cardiac puncture at 6 dpi. Blood was collected in serum separator tubes (BD), and serum was separated by centrifugation at 8,000 rpm for 5 min. Tissues and serum from infected animals were stored at −80°C until RNA isolation. Tissue samples and serum from ZIKV-infected mice were extracted with the RNeasy minikit (tissues) or viral RNA minikit (serum) (Qiagen). ZIKV RNA levels were determined by TaqMan one-step qRT-PCR on a CFX96 Touch real-time PCR detection system (Bio-Rad) under standard cycling conditions. The viral burden is expressed as genome copies per milliliter on a log10 scale after comparison with a standard curve produced using serial 100-fold dilutions of the ZIKV A plasmid. A previously reported primer set was used to detect ZIKV RNA: forward primer CCGCTGCCCAACACAAG, reverse primer CCACTAACGTTCTTTTGCAGACAT, and probe 5′-FAM (6-carboxyfluorescein)-AGCCTACCT-ZEN-TGACAAGCAATCAGACACTCAA-3′IABkFQ (Integrated DNA Technologies) (11, 50).
Deep sequencing.
Viruses were grown on Vero cells, virions were concentrated over a 20% sucrose cushion by ultracentrifugation, and full-length virion RNA was extracted using TRIzol (Ambion) according to the manufacturer’s protocol. PRVABC59 RNA was submitted to the UNC Chapel Hill Vironomics Core for library construction and sequencing on an Ion S5 instrument (Thermo Fisher Scientific). H/PF/2013 cDNA was made using extracted full-length virion RNA using random primer 9 (NEB) and SuperScript II, and the second strand of the cDNA was synthesized using the NEBNext Ultra II nondirectional RNA second-strand synthesis module (NEB) according to the manufacturer’s suggested protocol. Libraries were constructed from this double-stranded cDNA using a Nextera XT DNA library prep kit according to the manufacturer’s suggested protocol and quantified using a 4200 TapeStation system (Agilent Technologies) and a Qubit dsDNA HS assay kit (Thermo Fisher Scientific). Libraries were sequenced on a MiSeq desktop sequencer (Illumina).
Primer ID.
Viral RNA was extracted using a QIAamp viral RNA minikit (Qiagen). Primer ID sequencing libraries were constructed as previously described (51). In brief, we used the SuperScript III kit (Thermo Fisher) with cDNA primers containing a block of degenerate nucleotides (the Primer ID) to synthesize cDNA. After bead purification of the cDNA product using RNAClean XP (Beckman), two rounds of PCR were performed to incorporate MiSeq sequencing adaptors. Amplified products were purified, pooled, and sequenced using Illumina MiSeq 300-bp paired-end sequencing. A detailed protocol can be accessed at https://www.protocols.io/view/primer-id-miseq-library-prep-useewbe. We used the Illumina bcl2fastq pipeline for the initial data processing and the TCS pipeline (https://github.com/SwanstromLab/PID) to construct template consensus sequences.
Data analysis.
All data were analyzed with GraphPad Prism software. For viral burden analysis, the log-transformed titers were analyzed by a 2-tailed unpaired t test. Viral growth curves were compared by 2-way analysis of variance (ANOVA) with multiple comparisons. A P value of <0.05 indicated statistically significant differences.
ACKNOWLEDGMENTS
This work was supported by grants R01AI39512 and R21AI129431 to H.M.L. D.L.C. was supported by grant T32AI007419. This work also was supported by grants R01AI140970 to R.S. and R21AI137887 to N.J.M.
We acknowledge the efforts of the UNC High Throughput Sequencing Facility.
UNC is pursuing IP protection for Primer ID; R.S. is listed as a coinventor and has received nominal royalties.
REFERENCES
- 1.Petersen LR, Jamieson DJ, Powers AM, Honein MA. 2016. Zika virus. N Engl J Med 374:1552–1563. doi: 10.1056/NEJMra1602113. [DOI] [PubMed] [Google Scholar]
- 2.Pierson TC, Diamond MS. 2018. The emergence of Zika virus and its new clinical syndromes. Nature 560:573–581. doi: 10.1038/s41586-018-0446-y. [DOI] [PubMed] [Google Scholar]
- 3.Oehler E, Watrin L, Larre P, Leparc-Goffart I, Lastère S, Valour F, Baudouin L, Mallet HP, Musso D, Ghawche F. 2014. Zika virus infection complicated by Guillain-Barré syndrome—case report, French Polynesia, December 2013. Euro Surveill 19:20720. doi: 10.2807/1560-7917.es2014.19.9.20720. [DOI] [PubMed] [Google Scholar]
- 4.Coyne CB, Lazear HM. 2016. Zika virus—reigniting the TORCH. Nat Rev Microbiol 14:707–715. doi: 10.1038/nrmicro.2016.125. [DOI] [PubMed] [Google Scholar]
- 5.Brady OJ, Osgood-Zimmerman A, Kassebaum NJ, Ray SE, de Araújo VEM, da Nóbrega AA, Frutuoso LCV, Lecca RCR, Stevens A, Zoca de Oliveira B, de Lima JM Jr, Bogoch II, Mayaud P, Jaenisch T, Mokdad AH, Murray CJL, Hay SI, Reiner RC Jr, Marinho F. 2019. The association between Zika virus infection and microcephaly in Brazil 2015–2017: an observational analysis of over 4 million births. PLoS Med 16:e1002755. doi: 10.1371/journal.pmed.1002755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heang V, Yasuda CY, Sovann L, Haddow AD, da Rosa APT, Tesh RB, Kasper MR. 2012. Zika virus infection, Cambodia, 2010. Emerg Infect Dis 18:349–351. doi: 10.3201/eid1802.111224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Musso D, Bossin H, Mallet HP, Besnard M, Broult J, Baudouin L, Levi JE, Sabino EC, Ghawche F, Lanteri MC, Baud D. 2018. Zika virus in French Polynesia 2013–14: anatomy of a completed outbreak. Lancet Infect Dis 18:e172–e182. doi: 10.1016/S1473-3099(17)30446-2. [DOI] [PubMed] [Google Scholar]
- 8.Lanciotti RS, Lambert AJ, Holodniy M, Saavedra S, Del Carmen Castillo Signor L. 2016. Phylogeny of Zika virus in Western Hemisphere, 2015. Emerg Infect Dis 22:933–935. doi: 10.3201/eid2205.160065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsetsarkin KA, Kenney H, Chen R, Liu G, Manukyan H, Whitehead SS, Laassri M, Chumakov K, Pletnev AG. 2016. A full-length infectious cDNA clone of Zika virus from the 2015 epidemic in Brazil as a genetic platform for studies of virus-host interactions and vaccine development. mBio 7:e01114-16. doi: 10.1128/mBio.01114-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Widman DG, Young E, Yount BL, Plante KS, Gallichotte EN, Carbaugh DL, Peck KM, Plante J, Swanstrom J, Heise MT, Lazear HM, Baric RS. 2017. A reverse genetics platform that spans the Zika virus family tree. mBio 8:e02014-16. doi: 10.1128/mBio.02014-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carbaugh DL, Baric RS, Lazear HM. 2019. Envelope protein glycosylation mediates Zika virus pathogenesis. J Virol 93:e00113-19. doi: 10.1128/JVI.00113-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manangeeswaran M, Ireland DDC, Verthelyi D. 2016. Zika (PRVABC59) infection is associated with T cell infiltration and neurodegeneration in CNS of immunocompetent neonatal C57Bl/6 mice. PLoS Pathog 12:e1006004. doi: 10.1371/journal.ppat.1006004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goo L, Demaso CR, Pelc RS, Kuhn RJ, Pierson TC, Ledgerwood JE, Graham BS. 2018. The Zika virus envelope protein glycan loop regulates virion antigenicity. Virology 515:191–202. doi: 10.1016/j.virol.2017.12.032. [DOI] [PubMed] [Google Scholar]
- 14.Gong D, Zhang T-H, Zhao D, Du Y, Chapa TJ, Shi Y, Wang L, Contreras D, Zeng G, Shi P-Y, Wu T-T, Arumugaswami V, Sun R. 2018. High-throughput fitness profiling of Zika virus E protein reveals different roles for glycosylation during infection of mammalian and mosquito cells. iScience 1:97–111. doi: 10.1016/j.isci.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McDonald EM, Duggal NK, Delorey MJ, Oksanish J, Ritter JM, Brault AC. 2019. Duration of seminal Zika viral RNA shedding in immunocompetent mice inoculated with Asian and African genotype viruses. Virology 535:1–10. doi: 10.1016/j.virol.2019.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weger-Lucarelli J, Garcia SM, Rückert C, Byas A, O’Connor SL, Aliota MT, Friedrich TC, O’Connor DH, Ebel GD. 2018. Using barcoded Zika virus to assess virus population structure in vitro and in Aedes aegypti mosquitoes. Virology 521:138–148. doi: 10.1016/j.virol.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pardy RD, Valbon SF, Richer MJ. 2019. Running interference: interplay between Zika virus and the host interferon response. Cytokine 119:7–15. doi: 10.1016/j.cyto.2019.02.009. [DOI] [PubMed] [Google Scholar]
- 18.Singh PK, Guest J-M, Kanwar M, Boss J, Gao N, Juzych MS, Abrams GW, Yu F-S, Kumar A. 2017. Zika virus infects cells lining the blood-retinal barrier and causes chorioretinal atrophy in mouse eyes. JCI Insight 2:e92340. doi: 10.1172/jci.insight.92340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duggal NK, McDonald EM, Weger-Lucarelli J, Hawks SA, Ritter JM, Romo H, Ebel GD, Brault AC. 2019. Mutations present in a low-passage Zika virus isolate result in attenuated pathogenesis in mice. Virology 530:19–26. doi: 10.1016/j.virol.2019.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weger-Lucarelli J, Duggal NK, Bullard-Feibelman K, Veselinovic M, Romo H, Nguyen C, Rückert C, Brault AC, Bowen RA, Stenglein M, Geiss BJ, Ebel GD. 2017. Development and characterization of recombinant virus generated from a New World Zika virus infectious clone. J Virol 91:e01765-16. doi: 10.1128/JVI.01765-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shrivastava S, Puri V, Dilley KA, Ngouajio E, Shifflett J, Oldfield LM, Fedorova NB, Hu L, Williams T, Durbin A, Amedeo P, Rashid S, Shabman RS, Pickett BE. 2018. Whole genome sequencing, variant analysis, phylogenetics, and deep sequencing of Zika virus strains. Sci Rep 8:15843. doi: 10.1038/s41598-018-34147-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jabara CB, Jones CD, Roach J, Anderson JA, Swanstrom R. 2011. Accurate sampling and deep sequencing of the HIV-1 protease gene using a Primer ID. Proc Natl Acad Sci U S A 108:20166–20171. doi: 10.1073/pnas.1110064108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lazear HM, Govero J, Smith AM, Platt DJ, Fernandez E, Miner JJ, Diamond MS. 2016. A mouse model of Zika virus pathogenesis. Cell Host Microbe 19:720–730. doi: 10.1016/j.chom.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ávila-Pérez G, Nogales A, Park J-G, Márquez-Jurado S, Iborra FJ, Almazan F, Martínez-Sobrido L. 2019. A natural polymorphism in Zika virus NS2A protein responsible of virulence in mice. Sci Rep 9:19968. doi: 10.1038/s41598-019-56291-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tripathi S, Balasubramaniam VRMT, Brown JA, Mena I, Grant A, Bardina SV, Maringer K, Schwarz MC, Maestre AM, Sourisseau M, Albrecht RA, Krammer F, Evans MJ, Fernandez-Sesma A, Lim JK, García-Sastre A. 2017. A novel Zika virus mouse model reveals strain specific differences in virus pathogenesis and host inflammatory immune responses. PLoS Pathog 13:e1006258. doi: 10.1371/journal.ppat.1006258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rossi SL, Tesh RB, Azar SR, Muruato AE, Hanley KA, Auguste AJ, Langsjoen RM, Paessler S, Vasilakis N, Weaver SC. 2016. Characterization of a novel murine model to study Zika virus. Am J Trop Med Hyg 94:1362–1369. doi: 10.4269/ajtmh.16-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aliota MT, Caine EA, Walker EC, Larkin KE, Camacho E, Osorio JE. 2016. Characterization of lethal Zika virus infection in AG129 mice. PLoS Negl Trop Dis 10:e0004682. doi: 10.1371/journal.pntd.0004682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith DR, Sprague TR, Hollidge BS, Valdez SM, Padilla SL, Bellanca SA, Golden JW, Coyne SR, Kulesh DA, Miller LJ, Haddow AD, Koehler JW, Gromowski GD, Jarman RG, Alera MTP, Yoon IK, Buathong R, Lowen RG, Kane CD, Minogue TD, Bavari S, Tesh RB, Weaver SC, Linthicum KJ, Pitt ML, Nasar F. 2018. African and Asian Zika virus isolates display phenotypic differences both in vitro and in vivo. Am J Trop Med Hyg 98:432–444. doi: 10.4269/ajtmh.17-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morrison TE, Diamond MS. 2017. Animal models of Zika virus infection, pathogenesis, and immunity. J Virol 91:e00009-17. doi: 10.1128/JVI.00009-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ding Q, Gaska JM, Douam F, Wei L, Kim D, Balev M, Heller B, Ploss A. 2018. Species-specific disruption of STING-dependent antiviral cellular defenses by the Zika virus NS2B3 protease. Proc Natl Acad Sci U S A 115:E6310–E6318. doi: 10.1073/pnas.1803406115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gorman MJ, Caine EA, Zaitsev K, Begley MC, Weger-Lucarelli J, Uccellini MB, Tripathi S, Morrison J, Yount BL, Dinnon KH, Rückert C, Young MC, Zhu Z, Robertson SJ, McNally KL, Ye J, Cao B, Mysorekar IU, Ebel GD, Baric RS, Best SM, Artyomov MN, Garcia-Sastre A, Diamond MS. 2018. An immunocompetent mouse model of Zika virus infection. Cell Host Microbe 23:672–685.e6. doi: 10.1016/j.chom.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grant A, Ponia SS, Tripathi S, Balasubramaniam V, Miorin L, Sourisseau M, Schwarz MC, Sánchez-Seco MP, Evans MJ, Best SM, García-Sastre A. 2016. Zika virus targets human STAT2 to inhibit type I interferon signaling. Cell Host Microbe 19:882–890. doi: 10.1016/j.chom.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar A, Hou S, Airo AM, Limonta D, Mancinelli V, Branton W, Power C, Hobman TC. 2016. Zika virus inhibits type‐I interferon production and downstream signaling. EMBO Rep 17:1766–1775. doi: 10.15252/embr.201642627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dowall SD, Graham VA, Rayner E, Atkinson B, Hall G, Watson RJ, Bosworth A, Bonney LC, Kitchen S, Hewson R. 2016. A susceptible mouse model for Zika virus infection. PLoS Negl Trop Dis 10:e0004658. doi: 10.1371/journal.pntd.0004658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, Saucedo-Cuevas L, Regla-Nava JA, Chai G, Sheets N, Tang W, Terskikh AV, Shresta S, Gleeson JG. 2016. Zika virus infects neural progenitors in the adult mouse brain and alters proliferation. Cell Stem Cell 19:593–598. doi: 10.1016/j.stem.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roby JA, Setoh YX, Hall RA, Khromykh AA. 2015. Post-translational regulation and modifications of flavivirus structural proteins. J Gen Virol 96:1551–1569. doi: 10.1099/vir.0.000097. [DOI] [PubMed] [Google Scholar]
- 37.Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E, Jones CT, Mukhopadhyay S, Chipman PR, Strauss EG, Baker TS, Strauss JH. 2002. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108:717–725. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, Zhang W, Ogata S, Clements D, Strauss JH, Baker TS, Kuhn RJ, Rossmann MG. 2004. Conformational changes of the flavivirus E glycoprotein. Structure 12:1607–1618. doi: 10.1016/j.str.2004.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Slon Campos JL, Mongkolsapaya J, Screaton GR. 2018. The immune response against flaviviruses. Nat Immunol 19:1189–1198. doi: 10.1038/s41590-018-0210-3. [DOI] [PubMed] [Google Scholar]
- 40.Huber RG, Lim XN, Ng WC, Sim AYL, Poh HX, Shen Y, Lim SY, Sundstrom KB, Sun X, Aw JG, Too HK, Boey PH, Wilm A, Chawla T, Choy MM, Jiang L, de Sessions PF, Loh XJ, Alonso S, Hibberd M, Nagarajan N, Ooi EE, Bond PJ, Sessions OM, Wan Y. 2019. Structure mapping of dengue and Zika viruses reveals functional long-range interactions. Nat Commun 10:1408. doi: 10.1038/s41467-019-09391-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li P, Wei Y, Mei M, Tang L, Sun L, Huang W, Zhou J, Zou C, Zhang S, Qin CF, Jiang T, Dai J, Tan X, Zhang QC. 2018. Integrative analysis of Zika virus genome RNA structure reveals critical determinants of viral infectivity. Cell Host Microbe 24:875–886.e5. doi: 10.1016/j.chom.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 42.Gokhale NS, McIntyre ABR, McFadden MJ, Roder AE, Kennedy EM, Gandara JA, Hopcraft SE, Quicke KM, Vazquez C, Willer J, Ilkayeva OR, Law BA, Holley CL, Garcia-Blanco MA, Evans MJ, Suthar MS, Bradrick SS, Mason CE, Horner SM. 2016. N6-methyladenosine in Flaviviridae viral RNA genomes regulates infection. Cell Host Microbe 20:654–665. doi: 10.1016/j.chom.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slonchak A, Khromykh AA. 2018. Subgenomic flaviviral RNAs: what do we know after the first decade of research. Antiviral Res 159:13–25. doi: 10.1016/j.antiviral.2018.09.006. [DOI] [PubMed] [Google Scholar]
- 44.Göertz GP, Abbo SR, Fros JJ, Pijlman GP. 2018. Functional RNA during Zika virus infection. Virus Res 254:41–53. doi: 10.1016/j.virusres.2017.08.015. [DOI] [PubMed] [Google Scholar]
- 45.Baronti C, Piorkowski G, Charrel RN, Boubis L, Leparc-Goffart I, de Lamballerie X. 2014. Complete coding sequence of Zika virus from a French Polynesia outbreak in 2013. Genome Announc 2:e00500-14. doi: 10.1128/genomeA.00500-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sukupolvi-Petty S, Brien JD, Austin SK, Shrestha B, Swayne S, Kahle K, Doranz BJ, Johnson S, Pierson TC, Fremont DH, Diamond MS. 2013. Functional analysis of antibodies against dengue virus type 4 reveals strain-dependent epitope exposure that impacts neutralization and protection. J Virol 87:8826–8842. doi: 10.1128/JVI.01314-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brien JD, Lazear HM, Diamond MS. 2013. Propagation, quantification, detection, and storage of West Nile virus. Curr Protoc Microbiol 31:15D.3.1–15D.3.18. doi: 10.1002/9780471729259.mc15d03s31. [DOI] [PubMed] [Google Scholar]
- 48.Oliphant T, Nybakken GE, Engle M, Xu Q, Nelson CA, Sukupolvi-Petty S, Marri A, Lachmi B-E, Olshevsky U, Fremont DH, Pierson TC, Diamond MS. 2006. Antibody recognition and neutralization determinants on domains I and II of West Nile virus envelope protein. J Virol 80:12149–12159. doi: 10.1128/JVI.01732-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Messer WB, Yount B, Hacker KE, Donaldson EF, Huynh JP, de Silva AM, Baric RS. 2012. Development and characterization of a reverse genetic system for studying dengue virus serotype 3 strain variation and neutralization. PLoS Negl Trop Dis 6:e1486. doi: 10.1371/journal.pntd.0001486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lanciotti RS, Kosoy OL, Laven JJ, Velez JO, Lambert AJ, Johnson AJ, Stanfield SM, Duffy MR. 2008. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg Infect Dis 14:1232–1239. doi: 10.3201/eid1408.080287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keys JR, Zhou S, Anderson JA, Eron JJ, Rackoff LA, Jabara C, Swanstrom R. 2015. Primer ID informs next-generation sequencing platforms and reveals preexisting drug resistance mutations in the HIV-1 reverse transcriptase coding domain. AIDS Res Hum Retroviruses 31:658–668. doi: 10.1089/AID.2014.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]