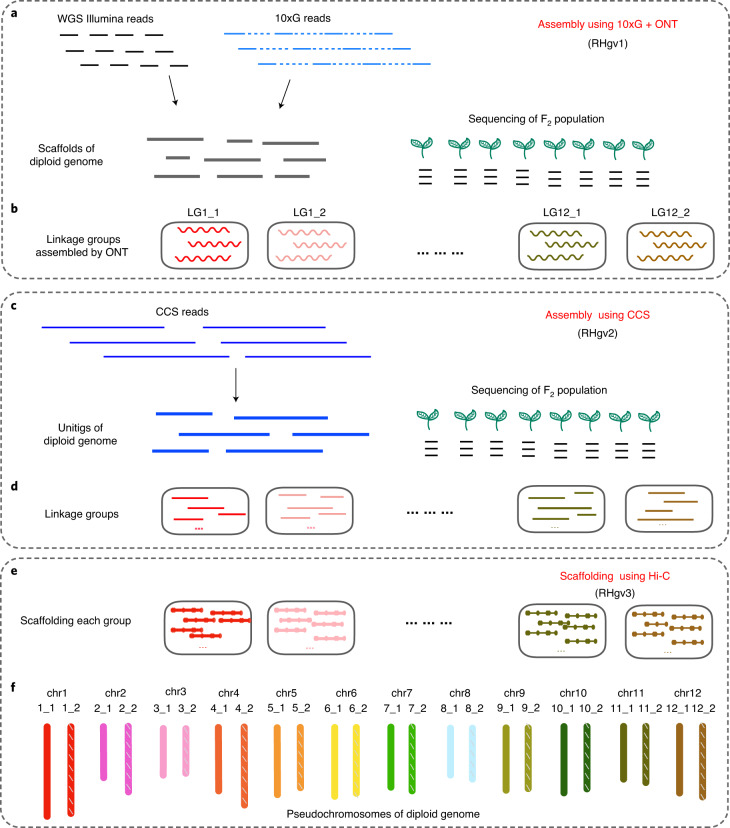
Fig. 1. Hybrid de novo assembly and phasing of the diploid potato genome.
a,b, The genome draft RHgv1 was assembled from WGS Illumina reads and 10xG linked reads, and the derived scaffolds were assigned into 24 haplotype-specific groups through genetic mapping based on a sequenced F2 population. The 24 groups represent chromosomes of the diploid potato (2n = 24). ONT reads were aligned to each linkage group and assembled to improve scaffold contiguity. c,d, A second genome sequence, RHgv2, was assembled from CCS reads. Similarly, unitigs were assigned into 24 groups through genetic mapping. e,f, The two assemblies were merged to generate a more comprehensive genome, RHgv3. Hi-C data were used to scaffold the sequences of each group into pseudochromosomes.