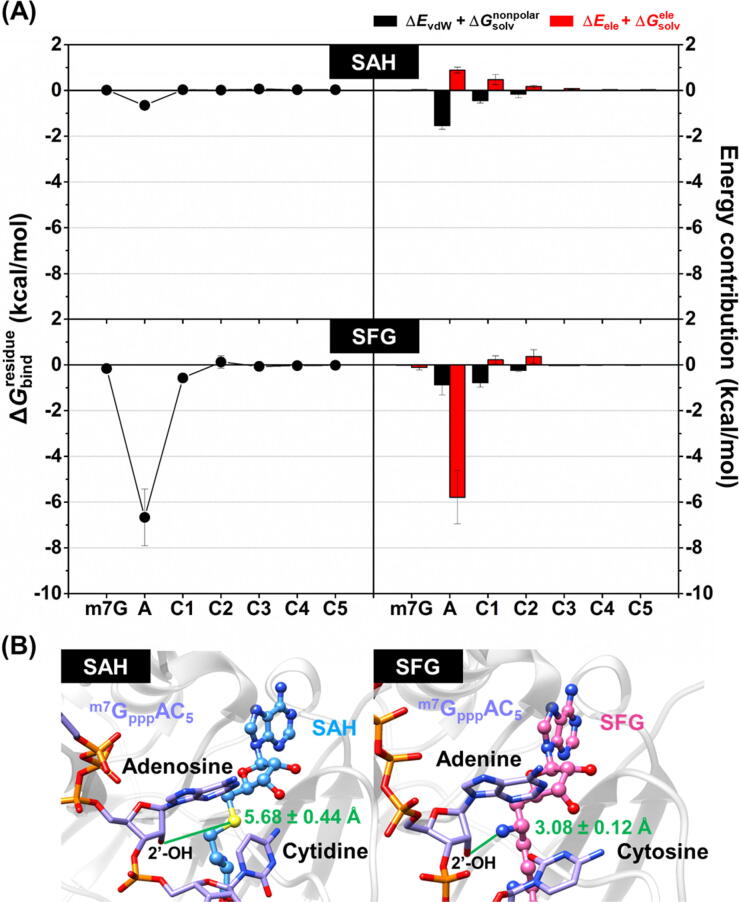
Fig. 5.
(A) (left) as well as electrostatic (ΔEele + , red) and vdW (ΔEvdW + , black) energy contribution from MD1 (right) of the m7GpppAC5 RNA substrate towards the binding of SAH (top) and SFG (bottom) to the SARS-CoV-2 nsp16/nsp10. Data are shown as mean ± SD of three independent simulations. (B) Representative 3D structures showing the distance (green line) between the sulfur atom of SAH (left) or the nitrogen atom of SFG (right) at 6′ position and the 2′-oxygen atom of RNA’s adenosine moiety in the enzyme active site. Distances are shown as mean ± SD (n = 3) calculated from the final snapshot of each complex. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)