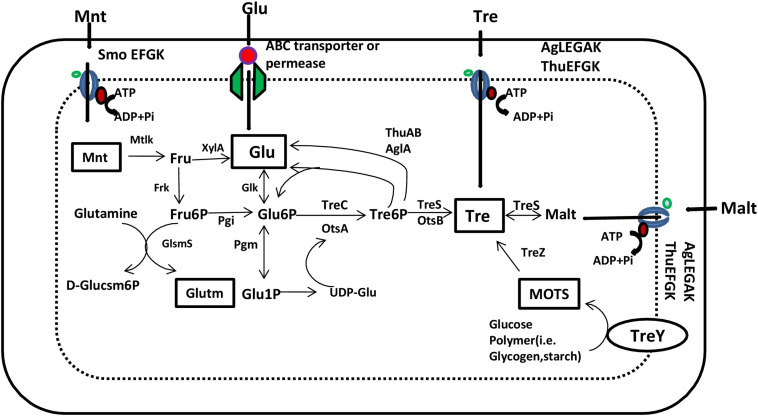
FIGURE 1.
Scheme of trehalose metabolism in Rhizobium etli based on the annotated genome. Glu, D-glucose; Glu6P, D-glucose-6-phosphate; Glu1P, D-glucose-1-phosphate; Glutm, D-glutamate; D-Glucsm6P, D-glucosamine-6-phosphate; Fru, D-fructose; Fru6P, D-fructose-6-phosphate; Malt, maltose; Mnt, mannitol; MOTS, maltoolygosyltrehalose; Tre, trehalose; TreP, trehalose-6-phosphate; AlgEFGAK and ThuEFGK, putative trehalose/maltose/sucrose ABC transporters; GlmS, glucosamine-6-phosphate synthase; Mtlk, mannitol 2-dehydrogenase; Frk, fructokinase; OtsA, trehalose-6-phosphate synthase; OtsB, trehalose-6-phosphate phosphatase; Pgi, phosphoglucose isomerase; XylA, xylose isomerase; TreC, trehalose-6-phosphate hydrolase; TreS, trehalose synthase; TreY, maltooligosyl trehalose synthase; TreZ, maltooligosyl trehalose trehalohydrolase; SmoEFGK, sorbitol/mannitol ABC transporter (source: Reina-Bueno et al., 2012).