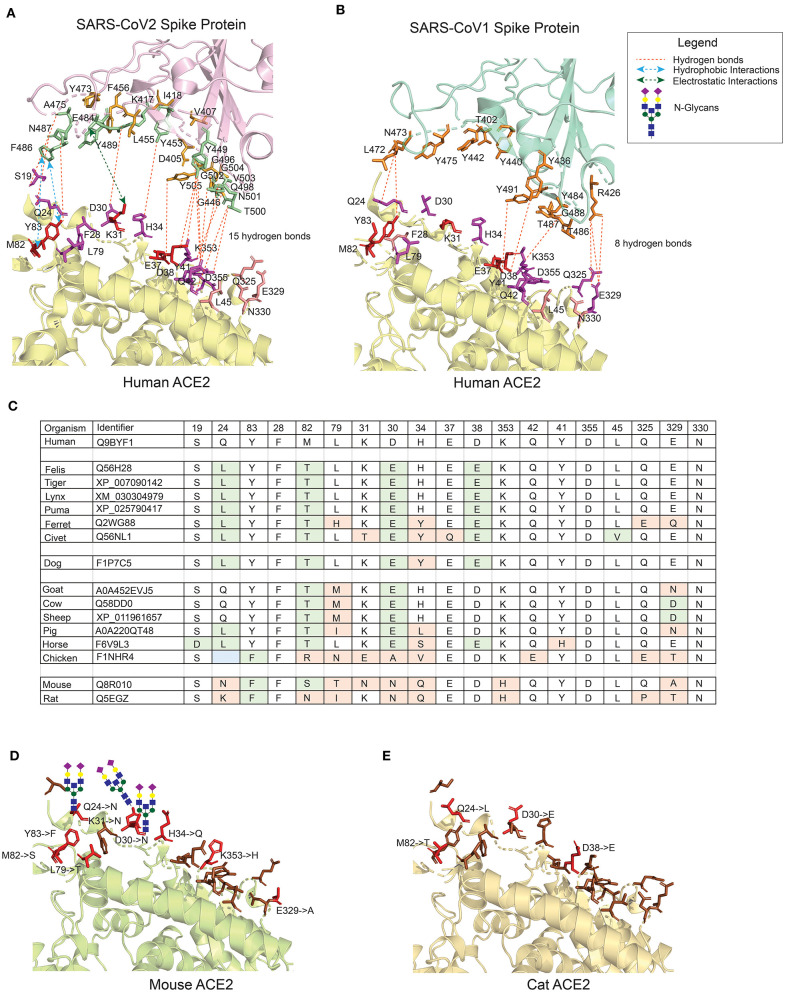
Figure 1.
The interface of ACE2 protein in different organisms with important amino-acid residues for interacting with the Spike (S) protein of SARS-CoV viruses. (A) The interacting interface between hACE2 protein and CoV2S (PDB ID: 6LZG). The residues in hACE2 proteins are colored in red, magenta and pink. Red residues are the most important and pink the least. The interacting residues in CoV2S are colored in green and orange colors where green residues are the important interacting residues. The distance between CoV2Spike and hACE2 proteins is increased to better visualize the residues and the interactions. (B) The interacting interface between hACE2 protein and CoV1S (PDB ID: 2AJF). Red residues on hACE2 protein are important residues for maintaining the interaction between hACE2 and CoV1S proteins. All the residues in CoV1S interacting with hACE2 are colored in orange. The distance between CoV1Spike and hACE2 proteins is increased to better visualize the residues and the interactions. Hydrogen bonds (A,B) as described in the structures of these interaction (PDB ID: 6LZG and 2AJF) and electrostatic and hydrophobic bonds in CoV1S-hACE2 interaction are depicted from Brielle et al. (4). (C) The positions mutated in ACE2 proteins in selected vertebrates that are either pets, domesticated or live in vicinity of humans. The mutated residues are shaded with green or orange background. The green background represents mutations resulting in similar property residue i.e. polar -> polar or non-polar -> non-polar. The orange background represents mutations resulting in changes in the residue property i.e. polar -> non-polar. (D) The interface of mouse's ACE2 protein. Red color residues represent the mutated residues. Three mutations on the left interacting region has resulted in an Arginine, introducing a glycan site. The basic structure of glycan is shown on the sites. (E) The interface of cat's ACE2 protein. The red color residues represent the mutated residues. The mouse's and cat's ACE2 structure are modelled with modeller-9.24 (30) using 6LZG as a template.